Step-by-Step Maturation Mechanism of Binary Toxin Pore Revealed by Cryo-EM Analysis
Yamada, T., Sugita, Y., Yoshida, T., Noda, T., Tsuge, H.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Iota toxin component Ib | 532 | Clostridium perfringens | Mutation(s): 0 | 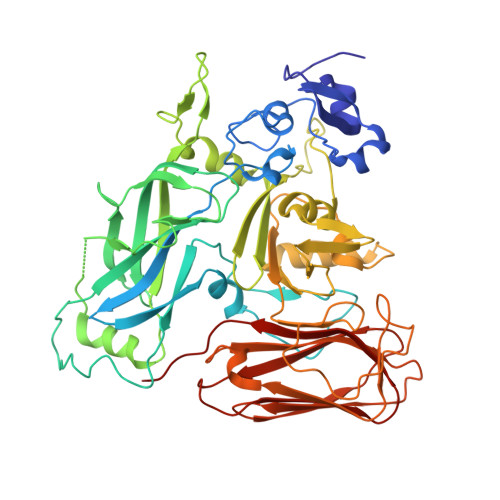 | |
UniProt | |||||
Find proteins for Q46221 (Clostridium perfringens) Explore Q46221 Go to UniProtKB: Q46221 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q46221 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CA (Subject of Investigation/LOI) Query on CA | B [auth A], C [auth A], D [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.19.2 |
| MODEL REFINEMENT | Coot | 0.9.5 |
| RECONSTRUCTION | cryoSPARC | 3.3.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | 21J13410 |
| Japan Society for the Promotion of Science (JSPS) | Japan | 21H02452 |
| Japan Society for the Promotion of Science (JSPS) | Japan | 24K01993 |
| Japan Agency for Medical Research and Development (AMED) | Japan | 2366 |