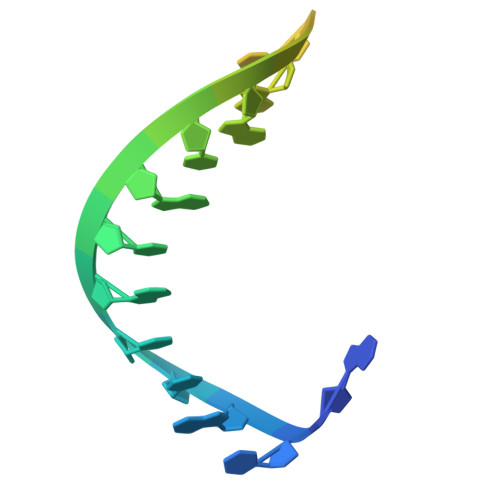
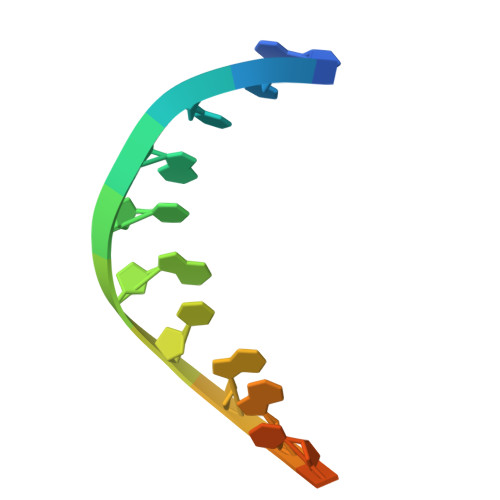
Target DNA-induced filament formation and nuclease activation of SPARDA complex.
Wang, F., Xu, H., Zhang, C., Xue, J., Li, Z.(2025) Cell Res 35: 510-519
- PubMed: 40122999
- DOI: https://doi.org/10.1038/s41422-025-01100-z
- Primary Citation of Related Structures:
9JSB, 9JSP, 9JSZ, 9JT2 - PubMed Abstract:
The short Argonaute-based bacterial defense system, SPARDA (Short Prokaryotic Argonaute and DNase/RNase-APAZ), utilizes guide RNA to target invading complementary DNA and exhibits collateral nuclease activity, leading to cell death or dormancy. However, its detailed mechanisms remain poorly understood. In this study, we investigated the SPARDA system from Novosphingopyxis baekryungensis (NbaSPARDA) and discovered an unexpected filament configuration upon target DNA binding, which strongly correlated with collateral nuclease activity. Filament formation and nuclease activation require a guide-target heteroduplex of sufficient length with perfect complementarity at the central region. A series of cryo-EM structures of NbaSPARDA complexes, loaded with guide RNA, target DNA of varying lengths, and substrate ssDNA, were determined at ~3.0 Å resolution. Structural analyses indicated that guide RNA binding induces dimerization of the NbaSPARDA complex, while target DNA engagement disrupts this dimerization. Further propagation of the guide-target heteroduplex triggers filament formation through a checkpoint mechanism. The NbaSPARDA filament consists of a backbone formed by interlocking short Argonaute proteins, with an inner layer composed of DREN nuclease domains. Filament formation leads to tetramerization of the monomeric DREN nuclease domain, activating its collateral nuclease activity against environmental nucleic acids - a feature leveraged for molecular diagnostics. For bacteria heterologously expressing the NbaSPARDA system, defense against invading bacteriophages and plasmids relies on filament formation. Collectively, these findings illustrate the detailed working mechanism of the NbaSPARDA complex and highlight the importance of its filament formation in host defense.
- State Key Laboratory of Biocatalysis and Enzyme Engineering, School of Life Sciences, Hubei University, Wuhan, Hubei, China.
Organizational Affiliation: