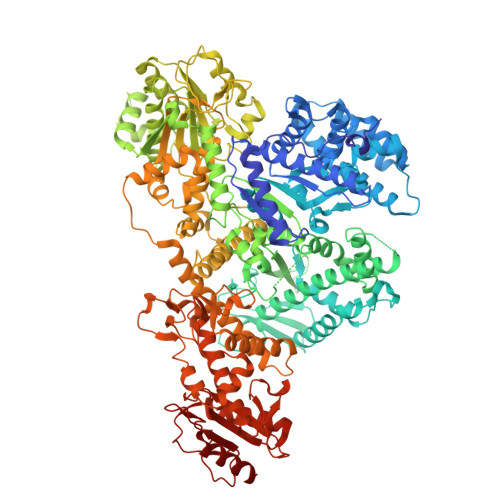
A gate-clamp mechanism for ssDNA translocation by DdmD in Vibrio cholerae plasmid defense.
Li, R., Liu, Y., Gao, H., Lin, Z.(2025) Nucleic Acids Res 53
- PubMed: 39907109
- DOI: https://doi.org/10.1093/nar/gkaf064
- Primary Citation of Related Structures:
9KHV, 9KHZ, 9KI0 - PubMed Abstract:
The DdmDE antiplasmid system, consisting of the helicase-nuclease DdmD and the prokaryotic Argonaute (pAgo) protein DdmE, plays a crucial role in defending Vibrio cholerae against plasmids. Guided by DNA, DdmE specifically targets plasmids, disassembles the DdmD dimer, and forms a DdmD-DdmE handover complex to facilitate plasmid degradation. However, the precise ATP-dependent DNA translocation mechanism of DdmD has remained unclear. Here, we present cryo-EM structures of DdmD bound to single-stranded DNA (ssDNA) in nucleotide-free, ATPγS-bound, and ADP-bound states. These structures, combined with biochemical analysis, reveal a unique "gate-clamp" mechanism for ssDNA translocation by DdmD. Upon ATP binding, arginine finger residues R855 and R858 reorient to interact with the γ-phosphate, triggering HD2 domain movement. This shift repositions the gate residue Q781, causing a flip of the 3' flank base, which is then clamped by residue F639. After ATP hydrolysis, the arginine finger releases the nucleotide, inducing HD2 to return to its open state. This conformational change enables DdmD to translocate along ssDNA by one nucleotide in the 5' to 3' direction. This study provides new insights into the ATP-dependent translocation of DdmD and contributes to understanding the mechanistic diversity within SF2 helicases.
Organizational Affiliation:
College of Chemical Engineering, Fuzhou University, Fujian 350108, China.