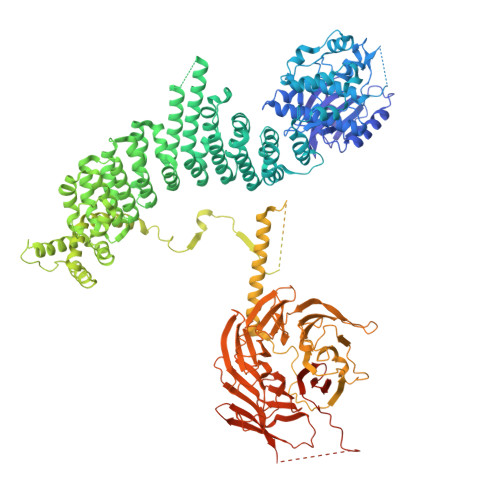
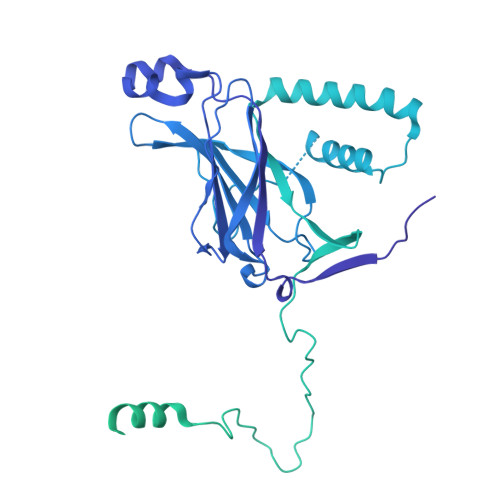
Structural pathway for PI3-kinase regulation by VPS15 in autophagy.
Cook, A.S.I., Chen, M., Nguyen, T.N., Cabezudo, A.C., Khuu, G., Rao, S., Garcia, S.N., Yang, M., Iavarone, A.T., Ren, X., Lazarou, M., Hummer, G., Hurley, J.H.(2025) Science 388: eadl3787-eadl3787
- PubMed: 39913640
- DOI: https://doi.org/10.1126/science.adl3787
- Primary Citation of Related Structures:
9MHF, 9MHG, 9MHH - PubMed Abstract:
The class III phosphatidylinositol-3 kinase complexes I and II (PI3KC3-C1 and PI3KC3-C2) have vital roles in macroautophagy and endosomal maturation, respectively. We elucidated a structural pathway of enzyme activation through cryo-electron microscopy analysis of PI3KC3-C1. The inactive conformation of the VPS15 pseudokinase stabilizes the inactive conformation, sequestering its N -myristate in the N-lobe of the pseudokinase. Upon activation, the myristate is liberated such that the VPS34 lipid kinase catalyzes phosphatidylinositol-3 phosphate production on membranes. The VPS15 pseudokinase domain binds tightly to guanosine triphosphate and stabilizes a web of interactions to autoinhibit the cytosolic complex and promote activation upon membrane binding. These findings show in atomistic detail how the VPS34 lipid kinase is activated in the context of a complete PI3K complex.
- Graduate Group in Biophysics, University of California, Berkeley, Berkeley, CA, USA.
Organizational Affiliation: