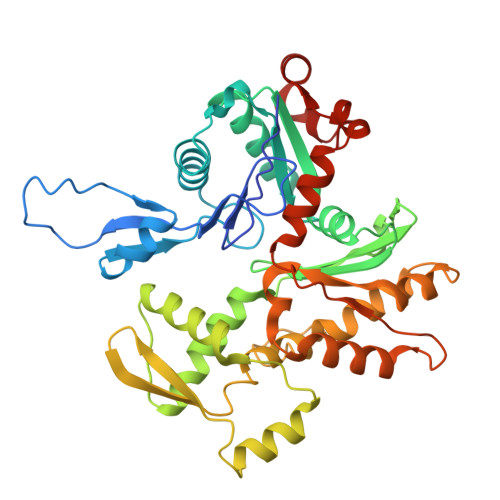
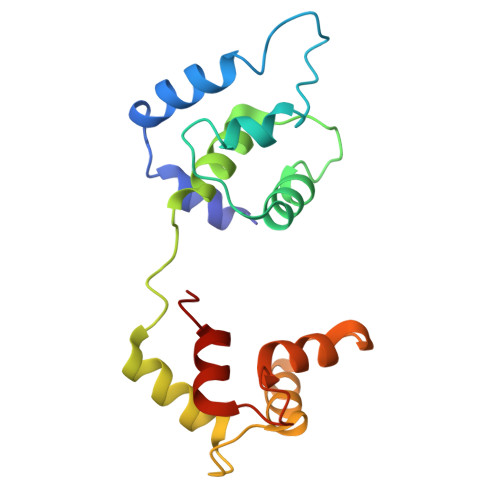
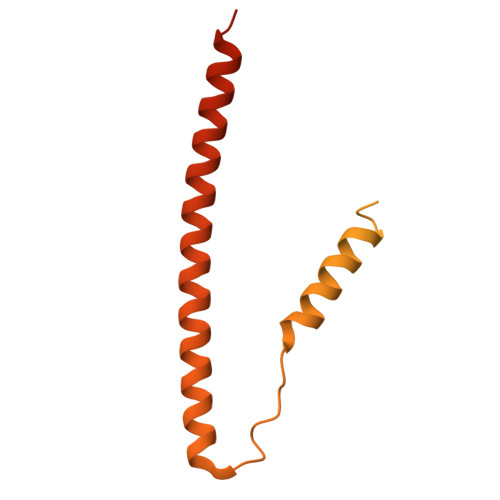
The role of the troponin T interactions with actin in regulation of cardiac thin filament revealed by the troponin T pathogenic variant Ile79Asn.
Risi, C.M., Landim-Vieira, M., Belknap, B., Chase, P.B., Pinto, J.R., Galkin, V.E.(2025) J Mol Cell Cardiol 204: 55-67
- PubMed: 40412797
- DOI: https://doi.org/10.1016/j.yjmcc.2025.05.005
- Primary Citation of Related Structures:
9E2E, 9MO4, 9MO5, 9MO6, 9MO7, 9MO8, 9MO9, 9MOA, 9MOB, 9MOC, 9MOD, 9MOI, 9MOK, 9MOL, 9MOM, 9MON, 9MOO, 9MOP, 9MOU, 9MOW, 9MOX - PubMed Abstract:
Cardiac muscle contraction/relaxation cycle depends on the rising and falling Ca 2+ levels in sarcomeres that control the extent of interactions between myosin-based thick and actin-based thin filaments. Cardiac thin filament (cTF) consists of actin, tropomyosin (Tm) that regulates myosin binding to actin, and troponin complex that governs Tm position upon Ca 2+ -binding. Troponin has three subunits - Ca 2+ -binding troponin C (TnC), Tm stabilizing troponin T (TnT), and inhibitory troponin I (TnI). TnT N-terminus (TnT1) interactions with actin stabilize the inhibited state of cTF. TnC, TnI, and Tm work in concert to control actomyosin interactions. Cryo-electron microscopy (cryo-EM) provided factual structures of healthy cTF, but structures of cTF carrying missense mutations linked to human cardiomyopathy are unknown. Variant Ile79Asn in human cardiac TnT (TnT-I79N) increases myofilament Ca 2+ sensitivity and slows cross-bridge kinetics, leading to severe hypertrophic/restrictive cardiomyopathy. Here, we used TnT-I79N mutation as a tool to examine the role of TnT1 in the complex mechanism of cTF regulation. Comparison of the cryo-EM structures of murine wild type and TnT-I79N native cTFs at systolic Ca 2+ levels (pCa = 5.8) demonstrates that TnT-I79N causes 1) dissociation of the TnT1 loop from its actin interface that results in Tm release to a more activated position, 2) reduced interaction of TnI C-terminus with actin-Tm, and 3) increased frequency of Ca 2+ -bound regulatory units. Our data indicate that the TnT1 loop is a crucial element of the allosteric regulatory network that couples Tn subunits and Tm to maintain adequate cTF response to physiological Ca 2+ levels during a heartbeat.
Organizational Affiliation:
Department of Biomedical and Translational Sciences, Macon & Joan Brock Virginia Health Sciences at Old Dominion University, Norfolk, VA 23507, USA.