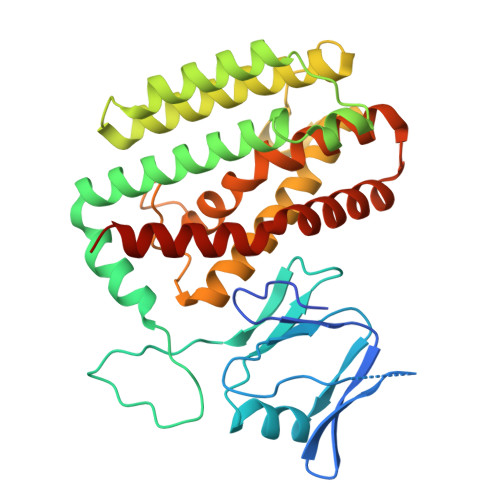
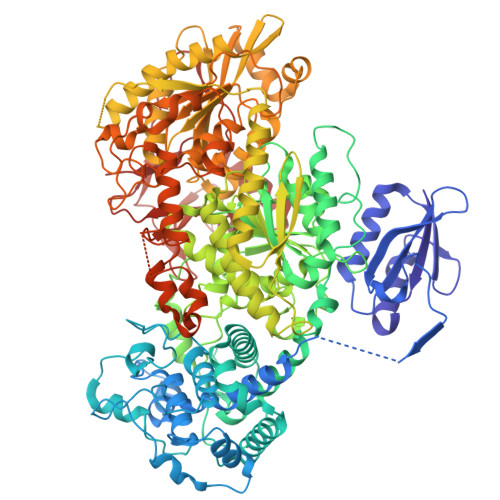
Structures reveal how the Cas1-2/3 integrase captures, delivers, and integrates foreign DNA into CRISPR loci.
Henriques, W.S., Bowman, J., Hall, L.N., Gauvin, C.C., Wei, H., Kuang, H., Zimanyi, C.M., Eng, E.T., Santiago-Frangos, A., Wiedenheft, B.(2025) Structure
- PubMed: 41072406
- DOI: https://doi.org/10.1016/j.str.2025.09.007
- Primary Citation of Related Structures:
9P11, 9P1D - PubMed Abstract:
Cas1 and Cas2 are the hallmark proteins of prokaryotic adaptive immunity. However, these two proteins are often fused to other proteins and the functional association of these fusions often remain poorly understood. Here we purify and determine structures of Cas1 and the Cas2/3 fusion proteins from Pseudomonas aeruginosa at distinct stages of CRISPR adaptation. Collectively, these structures reveal a prominent, positively charged channel on one face of the integration complex that captures short fragments of foreign DNA. Foreign DNA binding triggers conformational changes in Cas2/3 that expose new DNA binding surfaces necessary for homing the DNA-bound integrase to specific CRISPR loci. The length of the foreign DNA substrate determines if Cas1-2/3 docks completely onto the CRISPR repeat to successfully catalyze two sequential transesterification reactions required for integration. Together, these structures clarify how the Cas1-2/3 proteins orchestrate foreign DNA capture, site-specific delivery, and integration of new DNA into the bacterial genome.
- Department of Microbiology and Cell Biology, Montana State University, Bozeman, MT 59717, USA.
Organizational Affiliation: