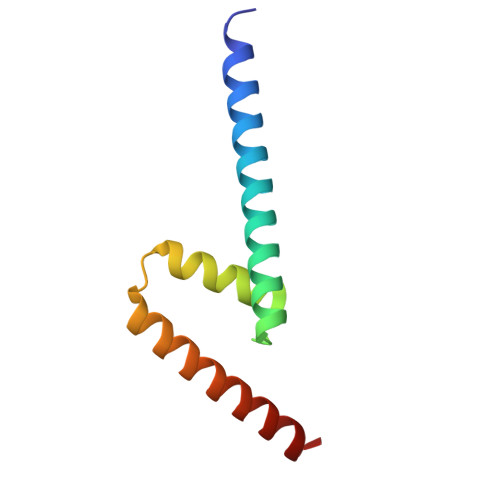
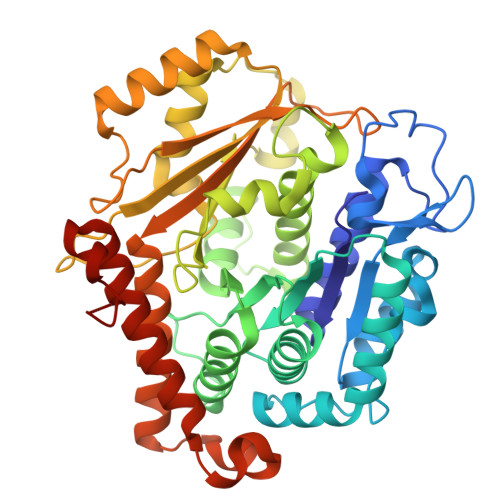
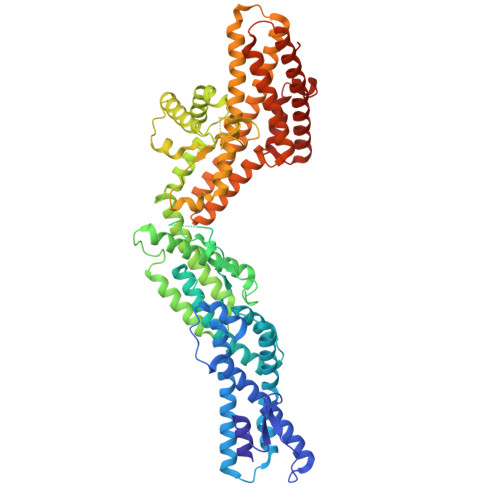
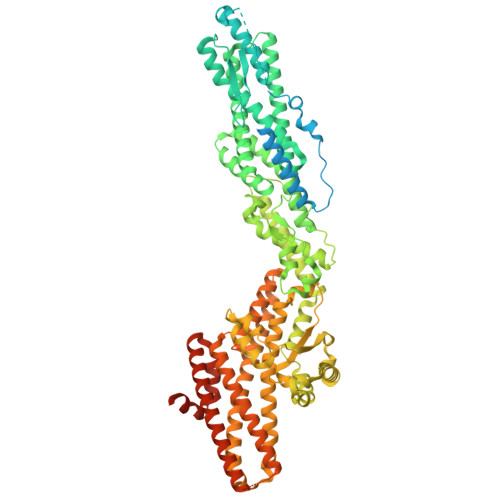
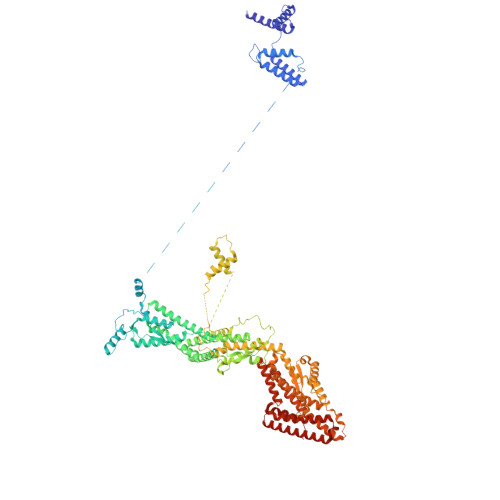
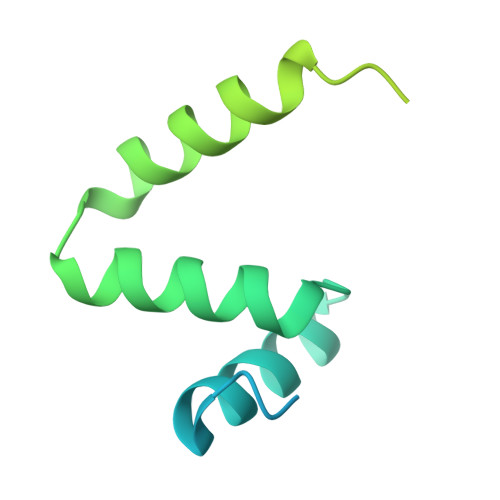
Structure of the microtubule-anchoring factor NEDD1 bound to the gamma-tubulin ring complex.
Munoz-Hernandez, H., Xu, Y., Pellicer Camardiel, A., Zhang, D., Xue, A., Aher, A., Walker, E., Marxer, F., Kapoor, T.M., Wieczorek, M.(2025) J Cell Biol 224
- PubMed: 40396914
- DOI: https://doi.org/10.1083/jcb.202410206
- Primary Citation of Related Structures:
9QVM, 9QVN - PubMed Abstract:
The γ-tubulin ring complex (γ-TuRC) is an essential multiprotein assembly that provides a template for microtubule nucleation. The γ-TuRC is recruited to microtubule-organizing centers (MTOCs) by the evolutionarily conserved attachment factor NEDD1. However, the structural basis of the NEDD1-γ-TuRC interaction is not known. Here, we report cryo-EM structures of NEDD1 bound to the human γ-TuRC in the absence or presence of the activating factor CDK5RAP2. We found that the C-terminus of NEDD1 forms a tetrameric α-helical assembly that contacts the lumen of the γ-TuRC cone and orients its microtubule-binding domain away from the complex. The structure of the γ-TuRC simultaneously bound to NEDD1 and CDK5RAP2 reveals that both factors can associate with the "open" conformation of the complex. Our results show that NEDD1 does not induce substantial conformational changes in the γ-TuRC but suggest that anchoring of γ-TuRC-capped microtubules by NEDD1 would be structurally compatible with the significant conformational changes experienced by the γ-TuRC during microtubule nucleation.
- Institute of Molecular Biology and Biophysics, ETH Zürich , Zürich, Switzerland.
Organizational Affiliation: