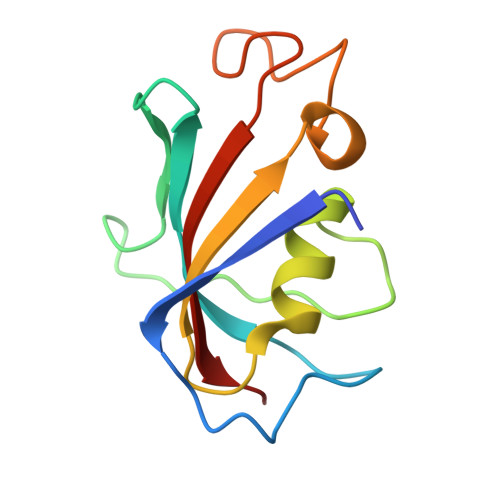
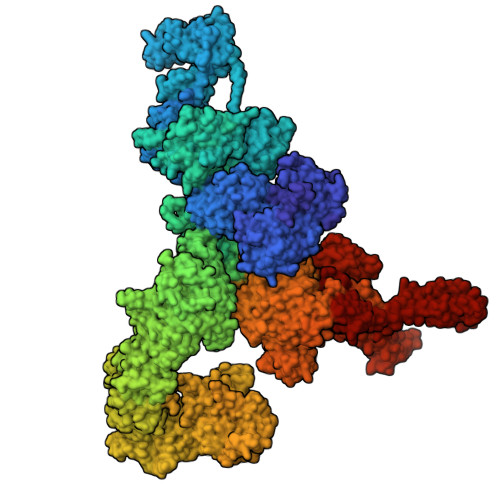
Cryo-EM investigation of ryanodine receptor type 3.
Chen, Y.S., Garcia-Castaneda, M., Charalambous, M., Rossi, D., Sorrentino, V., Van Petegem, F.(2024) Nat Commun 15: 8630-8630
- PubMed: 39366997
- DOI: https://doi.org/10.1038/s41467-024-52998-9
- Primary Citation of Related Structures:
9C1E, 9C1F - PubMed Abstract:
Ryanodine Receptor isoform 3 (RyR3) is a large ion channel found in the endoplasmic reticulum membrane of many different cell types. Within the hippocampal region of the brain, it is found in dendritic spines and regulates synaptic plasticity. It controls myogenic tone in arteries and is upregulated in skeletal muscle in early development. RyR3 has a unique functional profile with a very high sensitivity to activating ligands, enabling high gain in Ca 2+ -induced Ca 2+ release. Here we solve high-resolution cryo-EM structures of RyR3 in non-activating and activating conditions, revealing structural transitions that occur during channel opening. Addition of activating ligands yields only open channels, indicating an intrinsically high open probability under these conditions. RyR3 has reduced binding affinity to the auxiliary protein FKBP12.6 due to several sequence variations in the binding interface. We map disease-associated sequence variants and binding sites for known pharmacological agents. The N-terminal region contains ligand binding sites for a putative chloride anion and ATP, both of which are targeted by sequence variants linked to epileptic encephalopathy.
- Department of Biochemistry and Molecular Biology, the Life Sciences Centre, University of British Columbia, Vancouver, BC, Canada.
Organizational Affiliation: