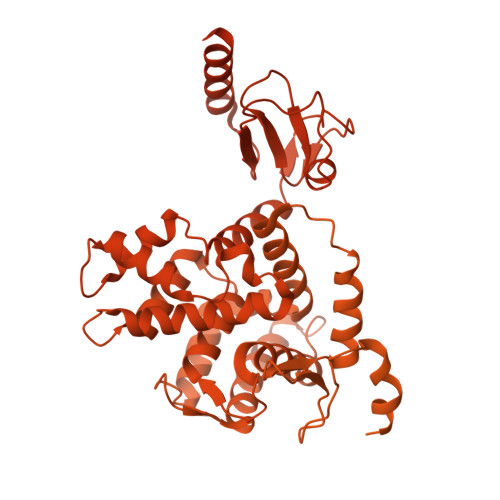
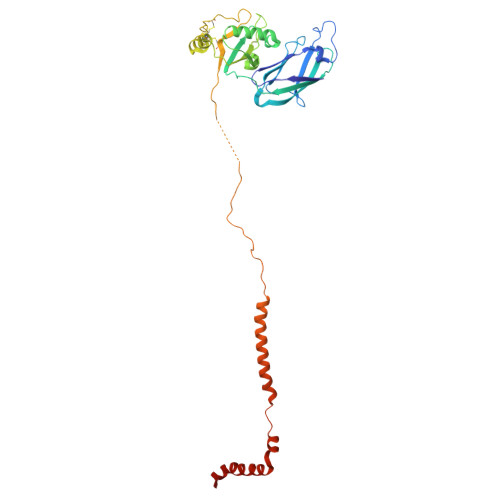
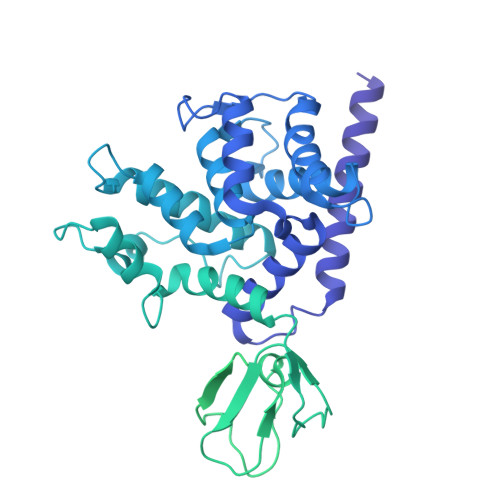
Native DGC structure rationalizes muscular dystrophy-causing mutations.
Liu, S., Su, T., Xia, X., Zhou, Z.H.(2025) Nature 637: 1261-1271
- PubMed: 39663457
- DOI: https://doi.org/10.1038/s41586-024-08324-w
- Primary Citation of Related Structures:
9C3C - PubMed Abstract:
Duchenne muscular dystrophy (DMD) is a severe X-linked recessive disorder marked by progressive muscle wasting leading to premature mortality 1,2 . Discovery of the DMD gene encoding dystrophin both revealed the cause of DMD and helped identify a family of at least ten dystrophin-associated proteins at the muscle cell membrane, collectively forming the dystrophin-glycoprotein complex (DGC) 3-9 . The DGC links the extracellular matrix to the cytoskeleton, but, despite its importance, its molecular architecture has remained elusive. Here we determined the native cryo-electron microscopy structure of rabbit DGC and conducted biochemical analyses to reveal its intricate molecular configuration. An unexpected β-helix comprising β-, γ- and δ-sarcoglycan forms an extracellular platform that interacts with α-dystroglycan, β-dystroglycan and α-sarcoglycan, allowing α-dystroglycan to contact the extracellular matrix. In the membrane, sarcospan anchors β-dystroglycan to the β-, γ- and δ-sarcoglycan trimer, while in the cytoplasm, β-dystroglycan's juxtamembrane fragment binds dystrophin's ZZ domain. Through these interactions, the DGC links laminin 2 to intracellular actin. Additionally, dystrophin's WW domain, along with its EF-hand 1 domain, interacts with α-dystrobrevin. A disease-causing mutation mapping to the WW domain weakens this interaction, as confirmed by deletion of the WW domain in biochemical assays. Our findings rationalize more than 110 mutations affecting single residues associated with various muscular dystrophy subtypes and contribute to ongoing therapeutic developments, including protein restoration, upregulation of compensatory genes and gene replacement.
- Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, Los Angeles, CA, USA.
Organizational Affiliation: