In vivo antibody diversification targeting a conserved coronavirus epitope.
Nair, U., Feng, Z., Akauliya, M., Esposito, A.G., Crain, C.R., Lamperti, E.D., Prum, T., Warner, J.E., Madungwe, L., Dale, G.A., Boucau, J., Gaiha, G.D., Yuan, M., Wilson, I.A., Batista, F.D.(2025) J Exp Medicine 222
- PubMed: 40674170
- DOI: https://doi.org/10.1084/jem.20241563
- Primary Citation of Related Structures:
9CPP, 9CPQ, 9CPR, 9CPS, 9CPT, 9CPU, 9CPV, 9CPW, 9CPX, 9CPY - PubMed Abstract:
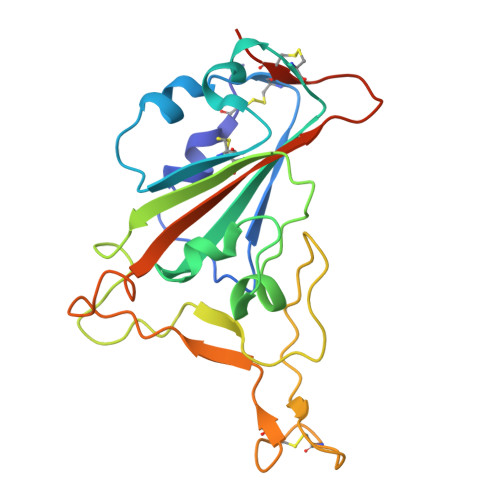
To explore the use of human B cell receptor (BCR) knock-in mice for broadening antibody responses, we diversified CR3022, a monoclonal antibody (mAb) originally identified in a 2003 severe acute respiratory syndrome coronavirus (SARS-CoV) convalescent patient. This mAb targets a conserved epitope on the coronavirus receptor-binding domain (RBD). We took advantage of high- and low-affinity CR3022 BCR knock-in mice and immunized them with SARS-CoV-2 Wuhan RBD trimers to expand the breadth of these antibodies toward this virus. The resulting antibodies retained the ability to neutralize SARS-CoV and exhibited enhanced affinity and neutralization against the SARS-CoV-2 WA1/2020 strain, as well as the Delta (B.1.617.2) and Omicron KP.3 variants. They also showed broadened reactivity to two bat coronaviruses: WIV1 and, to a lesser potency, BtKY72. Structural analysis revealed key mutations that enhanced binding and neutralization, highlighting the importance of epitope accessibility and variant-specific conformations in antibody diversification. These findings demonstrate that human BCR-expressing mouse models can generate effective antibodies with broad neutralizing activity against viral epitopes.
- The Ragon Institute of Mass General, MIT, and Harvard , Cambridge, MA, USA.
Organizational Affiliation: