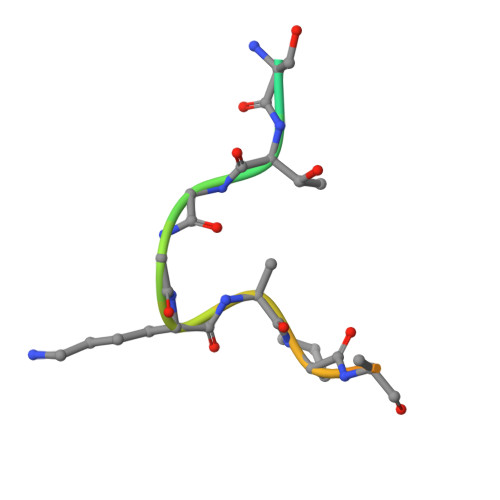
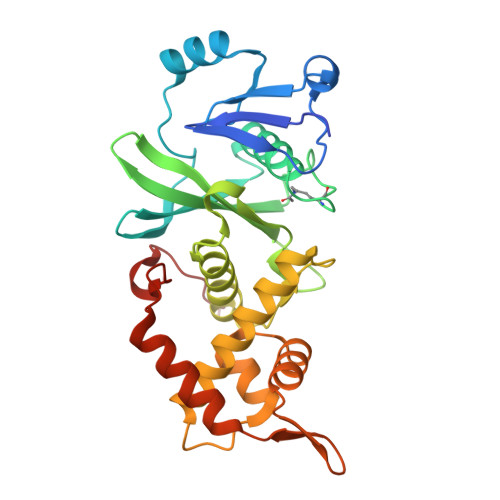
Modulation of the substrate preference of a MYST acetyltransferase by a scaffold protein.
Sengupta, R.N., Brodsky, O., Bingham, P., Diehl, W.C., Ferre, R., Greasley, S.E., Johnson, E., Kraus, M., Lieberman, W., Meier, J.L., Paul, T.A., Maegley, K.A.(2025) J Biological Chem 301: 108262-108262
- PubMed: 39909374
- DOI: https://doi.org/10.1016/j.jbc.2025.108262
- Primary Citation of Related Structures:
9DZN - PubMed Abstract:
The MYST family of lysine acetyltransferases are transcriptional regulators often dysregulated in cancer. In cells, MYST members form distinct multiprotein complexes that guide their histone substrate specificity, but how this selectivity is conferred is not fully understood. Here we interrogate a complex-mediated change in the substrate preference of the MYST member KAT6A, a target for cancer therapeutics. KAT6A forms a 4-protein complex with BRPF1, ING4/5, and MEAF6 to acetylate H3K23. However, additional substrates (H3K9, H3K14, and H3K27) have been proposed, and whether these residues are modified by KAT6A is unclear. We determined the histone substrate specificity of uncomplexed forms of KAT6A, including full-length KAT6A (KAT6A FL ) and the isolated acetyltransferase (MYST) domain, and the KAT6A FL 4-protein complex (KAT6A FL 4-plex). We show that the MYST domain and KAT6A FL preferentially acetylate H3K14, with this selectivity linked to a glycine pair preceding K14. A structure of the MYST domain bound to a H3K14-CoA bisubstrate inhibitor is consistent with a model in which the small size and flexibility of this glycine pair facilitates K14 acetylation. Notably, when KAT6A FL assembles into the 4-plex, H3K23 emerges as the favored substrate, with favorable recognition of an alanine-threonine pair before K23. These changes are mediated by BRPF1 and steady-state assays with H3 peptides indicate that this scaffold protein can alter the substrate preference of KAT6A FL by ≈10 3 -fold. Such context-dependent specificity illustrates how the functional properties of MYST members can be modulated by associated proteins and underscores the importance of characterizing these enzymes in their free and complexed forms.
Organizational Affiliation:
Oncology Research and Development, Pfizer, La Jolla, California, USA. Electronic address: raghuvir.sengupta@pfizer.com.