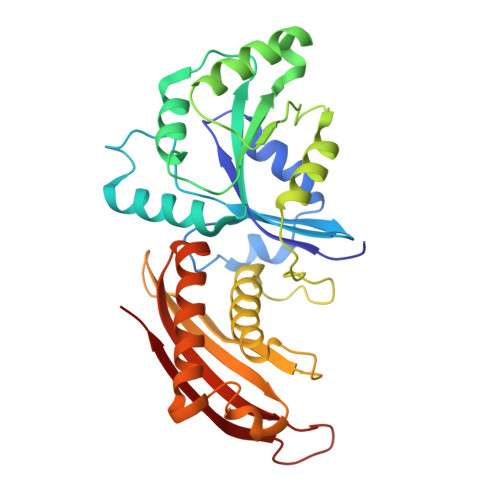
Crystal structure of the GDP-bound GTPase Era from Staphylococcus aureus.
Klochkova, E., Biktimirov, A., Islamov, D., Belousov, A., Validov, S., Yusupov, M., Usachev, K.(2024) Biochem Biophys Res Commun 735: 150852-150852
- PubMed: 39432921
- DOI: https://doi.org/10.1016/j.bbrc.2024.150852
- Primary Citation of Related Structures:
9JKP - PubMed Abstract:
GTPase Era from Staphylococcus aureus belongs to the TRAFAC superfamily of the TrmE-Era-EngA-EngB-Septin-like GTPases class and plays a significant role in the vital activity of this pathogenic microorganism as a maturation factor of the 30S ribosome subunit. However, the functions of this protein are not fully understood, making it a promising object for further study. Here, the 2.76 Å resolution crystal structure of Staphylococcus aureus Era in complex with GDP is presented. Structural comparison with other GTP-bound and GDP-bound homologous proteins, GTPase domain and the KH domain revealed a mutual orientation in S. aureus which has not been described before. The GDP-bound Era structure presented here will facilitate efforts to elucidate its interactions with its regulators and lay the foundation for a structure-based search for specific inhibitors.
- Kazan Federal University, 18 Kremlyovskaya St., 420008, Kazan, Russian Federation.
Organizational Affiliation: