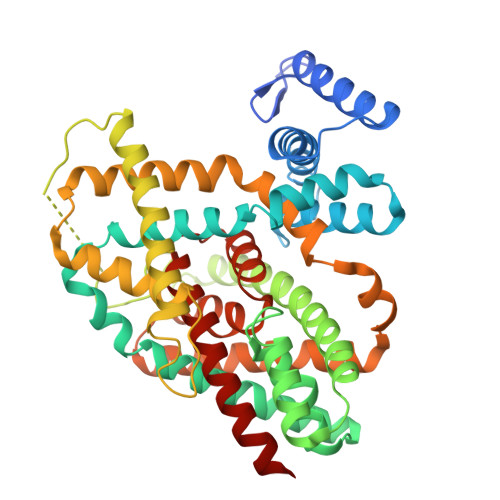
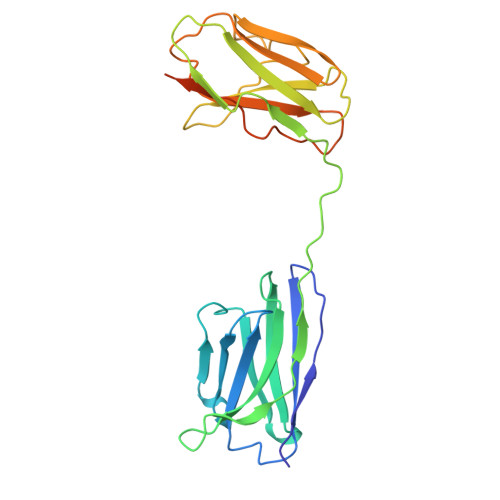
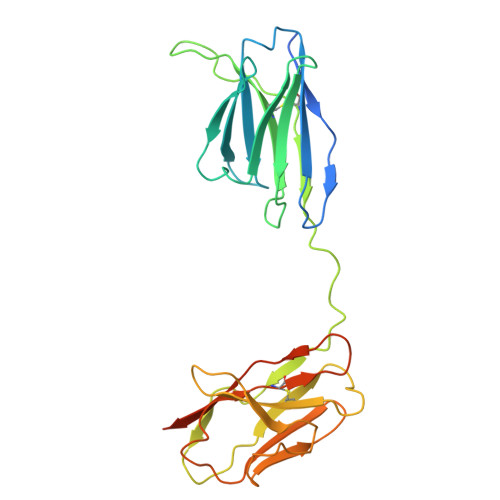
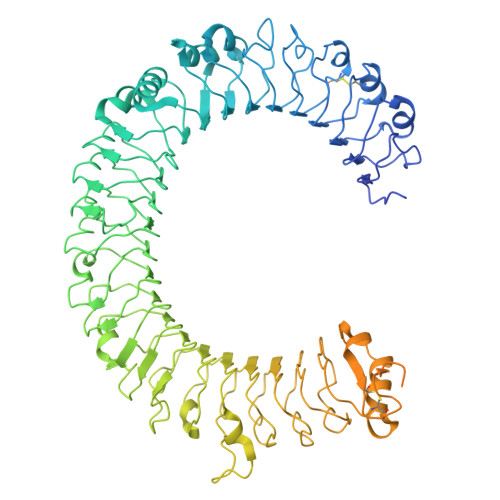
Disulfide-stabilized diabodies enable near-atomic cryo-EM imaging of small proteins: A case study of the bacterial Na + /citrate symporter CitS.
Kim, S., Kim, J.W., Park, J.G., Lee, S.S., Choi, S.H., Lee, J.O., Jin, M.S.(2025) Structure 33: 1088-1100.e3
- PubMed: 40169000
- DOI: https://doi.org/10.1016/j.str.2025.03.006
- Primary Citation of Related Structures:
9LSH, 9LSI, 9LSJ, 9LSK - PubMed Abstract:
Diabodies are engineered antibody fragments with two antigen-binding Fv domains. Previously, we demonstrated that they are often highly flexible but can be rigidified by introducing a disulfide bond at the Fv interface. In this study, we explored the potential of disulfide-bridged, bispecific diabodies for near-atomic cryoelectron microscopy (cryo-EM) imaging of small proteins because they can predictably link target proteins to "structural marker" proteins. As a case study, we used the bacterial citrate transporter CitS as the target protein, and the horseshoe-shaped ectodomain of human Toll-like receptor 3 (TLR3) as the marker. We show that diabodies containing one or two disulfide bonds enabled the 3D reconstruction of CitS at resolutions of 3.3 Å and 3.1 Å, respectively. This resolution surpassed previous crystallographic results and allowed us to visualize the high-resolution structural features of the transporter. Our work expands the application of diabodies in structural biology to address a key limitation in the field.
- Department of Life Sciences, Gwangju Institute of Science and Technology, Gwangju 61005, Republic of Korea.
Organizational Affiliation: