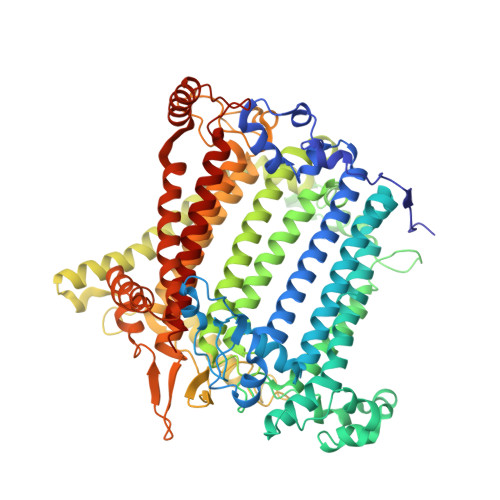
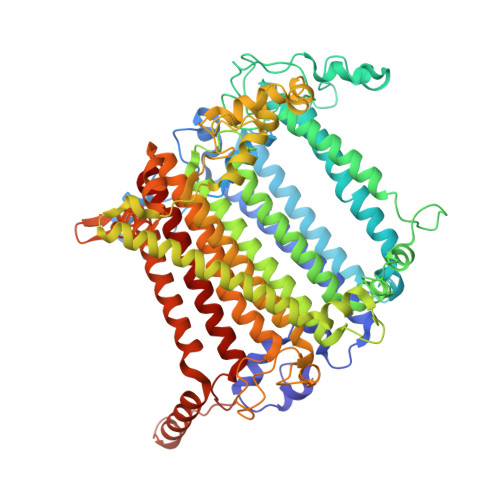
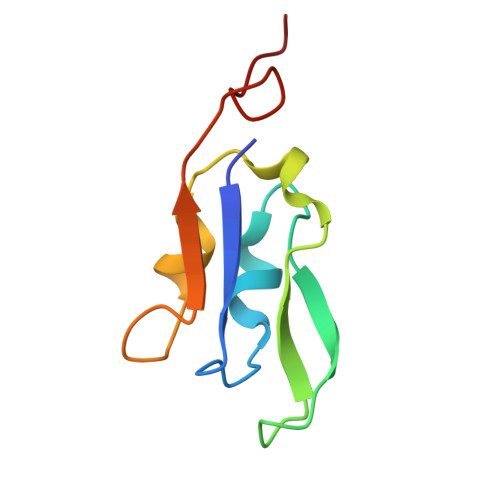
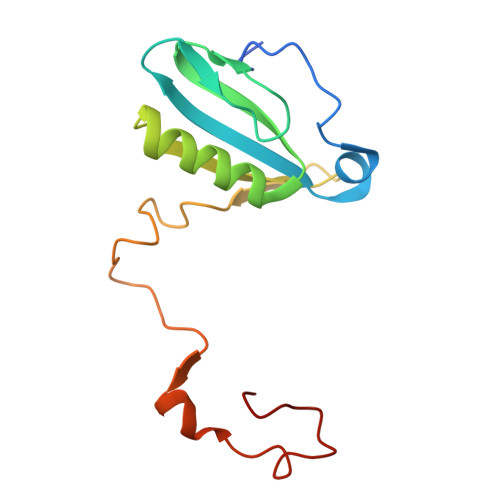
Architecture of photosystem I-light-harvesting complex from the eukaryotic filamentous yellow-green alga Tribonema minus.
Shao, R., Zou, Y., Shang, H., Qiu, Y., Liang, Z., Su, X., Zhang, S., Li, M., Pan, X.(2025) J Integr Plant Biol
- PubMed: 40778523
- DOI: https://doi.org/10.1111/jipb.70010
- Primary Citation of Related Structures:
9M4F - PubMed Abstract:
Eukaryotic photosystem I (PSI) is a multi-subunit pigment-protein supercomplex that consists of a core complex and multiple peripheral light-harvesting complexes I (LHCIs), which increases the light absorption capacity of the core complex. Throughout the evolution of oxygenic photoautotrophs, the core subunits of PSI have remained highly conserved, while LHCIs exhibit significant variability, presumably to adapt to diverse environments. This study presents a 2.82 Å resolution structure of PSI from the filamentous yellow-green alga Tribonema minus (Tm), a member of the class Xanthophyceae that evolved from red algae through endosymbiosis and is considered a promising candidate for biofuel production due to its high biomass and lipid content. Our structure reveals a supramolecular organization consisting of 12 core subunits and 13 LHCIs, here referred to as Xanthophyceae light-harvesting complexes (XLHs), along with the arrangement of pigments within the TmPSI-XLH supercomplex. A structural comparison between TmPSI-XLH and PSI-LHCI from various red lineages highlights distinctive features of TmPSI-XLH, suggesting that it represents a unique intermediate state in the PSI assembly process during the evolutionary transition from red algae to diatoms. Our findings advance the understanding of the molecular mechanisms responsible for energy transfer in Xanthophyceae PSI-XLH and the evolutionary adaptation of red lineages.
- Beijing Key Laboratory of Plant Gene Resources and Biotechnology for Carbon Reduction and Environmental Improvement, College of Life Science, Capital Normal University, Beijing, 100048, China.
Organizational Affiliation: