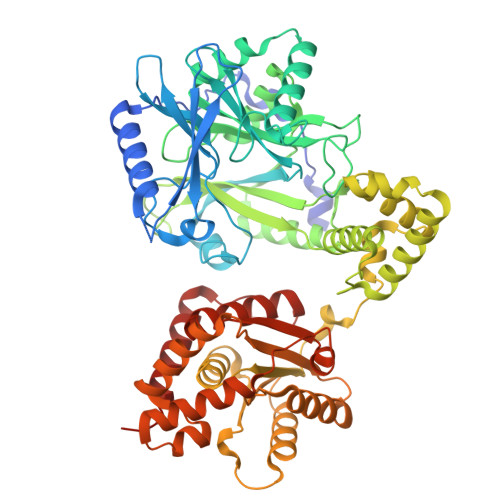
Interdomain assembly between the fungal tRNA ligase adenylyltransferase and kinase domain.
Kohler, S., Peschek, J.(2025) RNA
- PubMed: 40983467
- DOI: https://doi.org/10.1261/rna.080592.125
- Primary Citation of Related Structures:
9R3W - PubMed Abstract:
Trl1-type ligases play an essential role in fungi and plants during the non-conventional tRNA splicing as well as the unfolded protein response. The tripartite enzyme consists of an N-terminal adenylyltransferase domain (LIG), a central polynucleotide kinase domain (KIN) and a C-terminal cyclic phosphodiesterase domain (CPD). The Trl1-mediated reaction can be divided into two steps: (1) RNA end modification by the KIN and CPD domains, and (2) the adenylyltransferase reaction catalyzed by the LIG domain resulting in the phosphodiester bond formation. Due to its absence in humans, Trl1 is often discussed as potential target for antifungal therapy. To date structural information on the full-length Trl1 are missing. Several crystal structures of the individual LIG and KIN as well as a KIN-CPD construct have been solved, thereby elucidating the fold of the individual domains, their cofactor and substrate binding. Here, we provide the missing crystal structure of the two-domain LIG-KIN construct from the thermophilic fungus Chaetomium thermophilum revealing the interdomain assembly and interface. Based on our structure and complementing AlphaFold3 predictions, we further propose a model with implications for interdomain RNA substrate transfer.
- Universitat Heidelberg.
Organizational Affiliation: