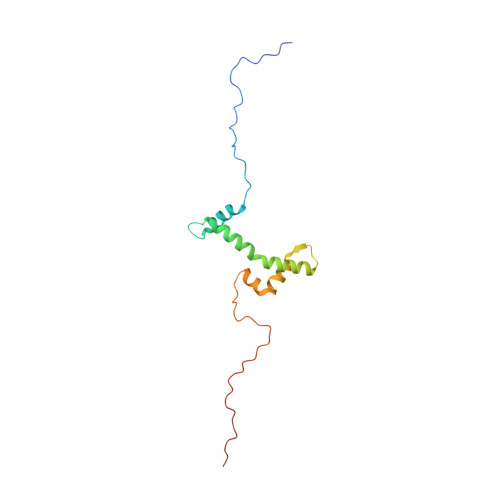
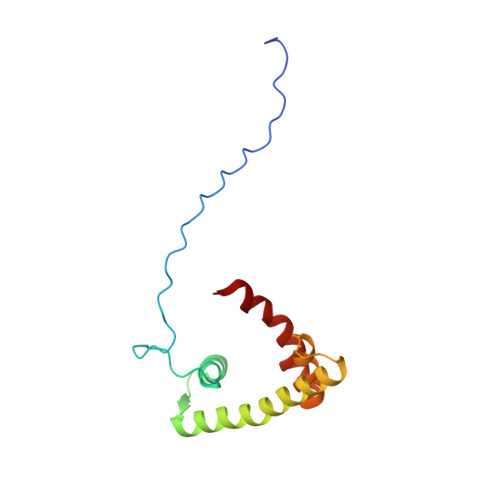
Solution structure of the isolated histone H2A-H2B heterodimer
Moriwaki, Y., Yamane, T., Ohtomo, H., Ikeguchi, M., Kurita, J., Sato, M., Nagadoi, A., Shimojo, H., Nishimura, Y.(2016) Sci Rep 6: 24999-24999
- PubMed: 27181506
- DOI: https://doi.org/10.1038/srep24999
- Primary Citation of Related Structures:
2RVQ - PubMed Abstract:
During chromatin-regulated processes, the histone H2A-H2B heterodimer functions dynamically in and out of the nucleosome. Although detailed crystal structures of nucleosomes have been established, that of the isolated full-length H2A-H2B heterodimer has remained elusive. Here, we have determined the solution structure of human H2A-H2B by NMR coupled with CS-Rosetta. H2A and H2B each contain a histone fold, comprising four α-helices and two β-strands (α1-β1-α2-β2-α3-αC), together with the long disordered N- and C-terminal H2A tails and the long N-terminal H2B tail. The N-terminal αN helix, C-terminal β3 strand, and 310 helix of H2A observed in the H2A-H2B nucleosome structure are disordered in isolated H2A-H2B. In addition, the H2A α1 and H2B αC helices are not well fixed in the heterodimer, and the H2A and H2B tails are not completely random coils. Comparison of hydrogen-deuterium exchange, fast hydrogen exchange, and {(1)H}-(15)N hetero-nuclear NOE data with the CS-Rosetta structure indicates that there is some conformation in the H2A 310 helical and H2B Lys11 regions, while the repression domain of H2B (residues 27-34) exhibits an extended string-like structure. This first structure of the isolated H2A-H2B heterodimer provides insight into its dynamic functions in chromatin.
Organizational Affiliation:
Graduate School of Medical Life Science, Yokohama City University, 1-7-29 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan.