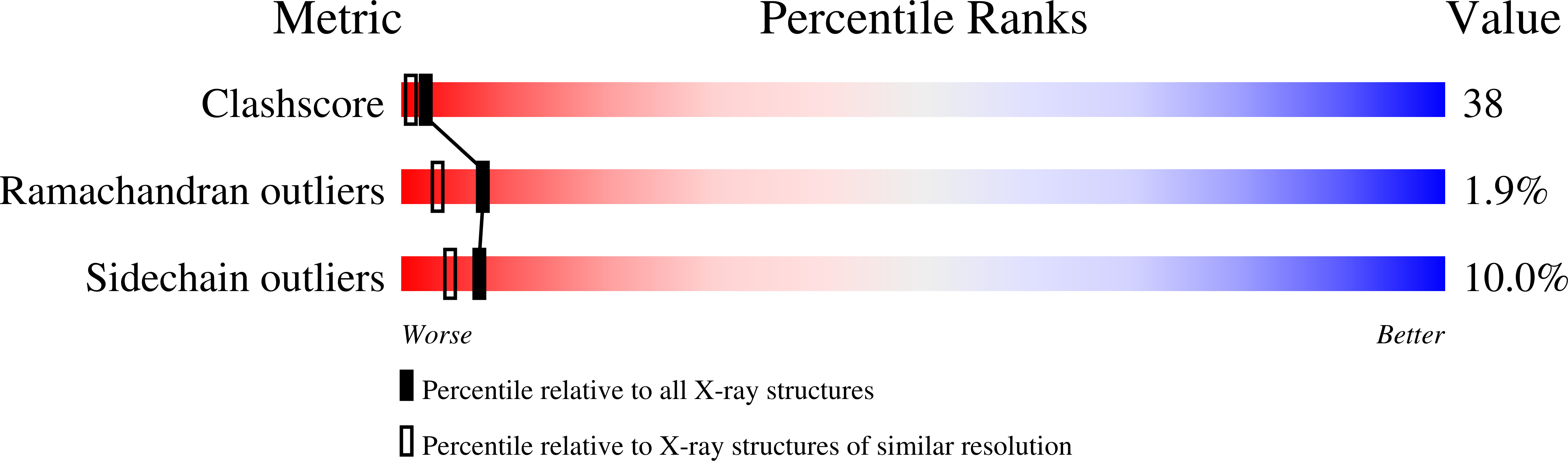Anticooperative ligand binding properties of recombinant ferric Vitreoscilla homodimeric hemoglobin: a thermodynamic, kinetic and X-ray crystallographic study.
Bolognesi, M., Boffi, A., Coletta, M., Mozzarelli, A., Pesce, A., Tarricone, C., Ascenzi, P.(1999) J Mol Biol 291: 637-650
- PubMed: 10448042
- DOI: https://doi.org/10.1006/jmbi.1999.2975
- Primary Citation of Related Structures:
3VHB, 4VHB - PubMed Abstract:
Thermodynamics and kinetics for cyanide, azide, thiocyanate and imidazole binding to recombinant ferric Vitreoscilla sp. homodimeric hemoglobin (Vitreoscilla Hb) have been determined at pH 6.4 and 7.0, and 20.0 degrees C, in solution and in the crystalline state. Moreover, the three-dimensional structures of the diligated thiocyanate and imidazole derivatives of recombinant ferric Vitreoscilla Hb have been determined by X-ray crystallography at 1.8 A (Rfactor=19.9%) and 2.1 A (Rfactor=23.8%) resolution, respectively. Ferric Vitreoscilla Hb displays an anticooperative ligand binding behaviour in solution. This very unusual feature can only be accounted for by assuming ligand-linked conformational changes in the monoligated species, which lead to the observed 300-fold decrease in the affinity of cyanide, azide, thiocyanate and imidazole for the monoligated ferric Vitreoscilla Hb with respect to that of the fully unligated homodimer. In the crystalline state, thermodynamics for azide and imidazole binding to ferric Vitreoscilla Hb may be described as a simple process with an overall ligand affinity for the homodimer corresponding to that for diligation in solution. These data suggest that the ligand-free homodimer, observed in the crystalline state, is constrained in a low affinity conformation whose ligand binding properties closely resemble those of the monoligated species in solution. From the kinetic viewpoint, anticooperativity is reflected by the 300-fold decrease of the second-order rate constant for cyanide and imidazole binding to the monoligated ferric Vitreoscilla Hb with respect to that for ligand association to the ligand-free homodimer in solution. On the other hand, values of the first-order rate constant for cyanide and imidazole dissociation from the diligated and monoligated derivatives of ferric Vitreoscilla Hb in solution are closely similar. As a whole, ligand binding and structural properties of ferric Vitreoscilla Hb appear to be unique among all Hbs investigated to date.
Organizational Affiliation:
Centro per le Biotecnologie Avanzate-IST and Dipartimento di Fisica-INFM, Largo Rosanna Benzi 10, Università di Genova, I-16132, Italy.