Convergent immunological solutions to Argentine hemorrhagic fever virus neutralization.
Zeltina, A., Krumm, S.A., Sahin, M., Struwe, W.B., Harlos, K., Nunberg, J.H., Crispin, M., Pinschewer, D.D., Doores, K.J., Bowden, T.A.(2017) Proc Natl Acad Sci U S A 114: 7031-7036
- PubMed: 28630325
- DOI: https://doi.org/10.1073/pnas.1702127114
- Primary Citation of Related Structures:
5NUZ - PubMed Abstract:
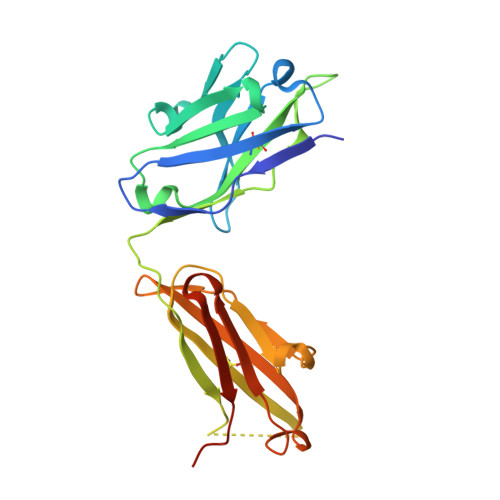
Transmission of hemorrhagic fever New World arenaviruses from their rodent reservoirs to human populations poses substantial public health and economic dangers. These zoonotic events are enabled by the specific interaction between the New World arenaviral attachment glycoprotein, GP1, and cell surface human transferrin receptor (hTfR1). Here, we present the structural basis for how a mouse-derived neutralizing antibody (nAb), OD01, disrupts this interaction by targeting the receptor-binding surface of the GP1 glycoprotein from Junín virus (JUNV), a hemorrhagic fever arenavirus endemic in central Argentina. Comparison of our structure with that of a previously reported nAb complex (JUNV GP1-GD01) reveals largely overlapping epitopes but highly distinct antibody-binding modes. Despite differences in GP1 recognition, we find that both antibodies present a key tyrosine residue, albeit on different chains, that inserts into a central pocket on JUNV GP1 and effectively mimics the contacts made by the host TfR1. These data provide a molecular-level description of how antibodies derived from different germline origins arrive at equivalent immunological solutions to virus neutralization.
Organizational Affiliation:
Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, United Kingdom.