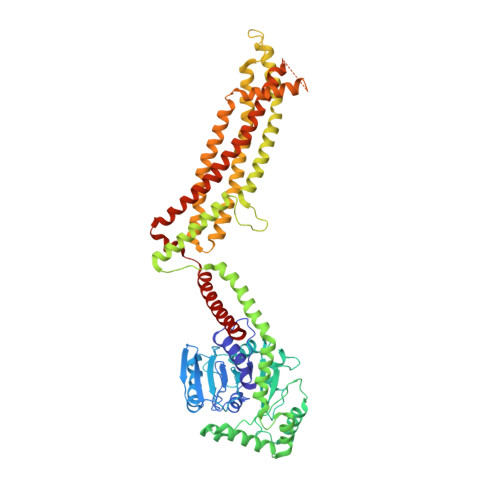
CryoEM structure of MxB reveals a novel oligomerization interface critical for HIV restriction.
Alvarez, F.J.D., He, S., Perilla, J.R., Jang, S., Schulten, K., Engelman, A.N., Scheres, S.H.W., Zhang, P.(2017) Sci Adv 3: e1701264-e1701264
- PubMed: 28929138
- DOI: https://doi.org/10.1126/sciadv.1701264
- Primary Citation of Related Structures:
5UOT - PubMed Abstract:
Human dynamin-like, interferon-induced myxovirus resistance 2 (Mx2 or MxB) is a potent HIV-1 inhibitor. Antiviral activity requires both the amino-terminal region of MxB and protein oligomerization, each of which has eluded structural determination due to difficulties in protein preparation. We report that maltose binding protein-fused, full-length wild-type MxB purifies as oligomers and further self-assembles into helical arrays in physiological salt. Guanosine triphosphate (GTP), but not guanosine diphosphate, binding results in array disassembly, whereas subsequent GTP hydrolysis allows its reformation. Using cryo-electron microscopy (cryoEM), we determined the MxB assembly structure at 4.6 Å resolution, representing the first near-atomic resolution structure in the mammalian dynamin superfamily. The structure revealed previously described and novel MxB assembly interfaces. Mutational analyses demonstrated a critical role for one of the novel interfaces in HIV-1 restriction.
Organizational Affiliation:
Department of Structural Biology, University of Pittsburgh School of Medicine, Pittsburgh, PA 15260, USA.