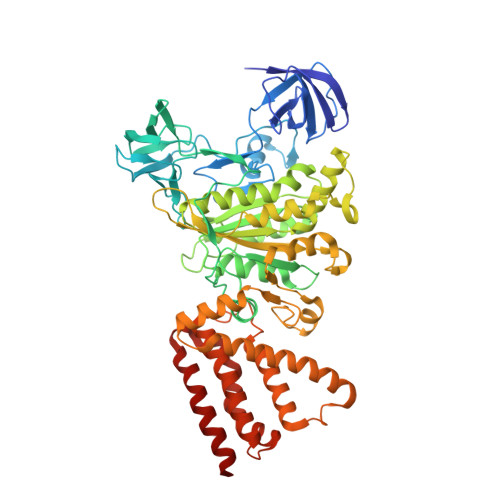
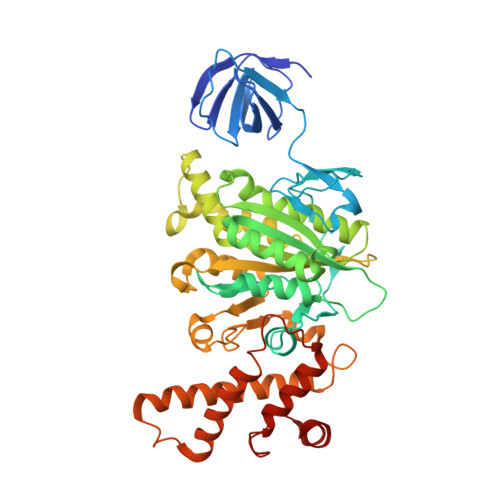
Metastable asymmetrical structure of a shaftless V1motor.
Maruyama, S., Suzuki, K., Imamura, M., Sasaki, H., Matsunami, H., Mizutani, K., Saito, Y., Imai, F.L., Ishizuka-Katsura, Y., Kimura-Someya, T., Shirouzu, M., Uchihashi, T., Ando, T., Yamato, I., Murata, T.(2019) Sci Adv 5: eaau8149-eaau8149
- PubMed: 30729160
- DOI: https://doi.org/10.1126/sciadv.aau8149
- Primary Citation of Related Structures:
5ZE9, 5ZEA - PubMed Abstract:
V 1 -ATPase is an ATP-driven rotary motor that is composed of a ring-shaped A 3 B 3 complex and a central DF shaft. The nucleotide-free A 3 B 3 complex of Enterococcus hirae , composed of three identical A 1 B 1 heterodimers, showed a unique asymmetrical structure, probably due to the strong binding of the N-terminal barrel domain, which forms a crown structure. Here, we mutated the barrel region to weaken the crown, and performed structural analyses using high-speed atomic force microscopy and x-ray crystallography of the mutant A 3 B 3 . The nucleotide-free mutant A 3 B 3 complex had a more symmetrical open structure than the wild type. Binding of nucleotides produced a closely packed spiral-like structure with a disrupted crown. These findings suggest that wild-type A 3 B 3 forms a metastable (stressed) asymmetric structure composed of unstable A 1 B 1 conformers due to the strong constraint of the crown. The results further the understanding of the principle of the cooperative transition mechanism of rotary motors.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Chiba University, 1-33 Yayoi-cho, Inage, Chiba 263-8522, Japan.