Chemoenzymatic Synthesis and Biological Evaluation for Bioactive Molecules Derived from Bacterial Benzoyl Coenzyme A Ligase and Plant Type III Polyketide Synthase.
Adhikari, K., Lo, I.W., Chen, C.L., Wang, Y.L., Lin, K.H., Zadeh, S.M., Rattinam, R., Li, Y.S., Wu, C.J., Li, T.L.(2020) Biomolecules 10
- PubMed: 32397467
- DOI: https://doi.org/10.3390/biom10050738
- Primary Citation of Related Structures:
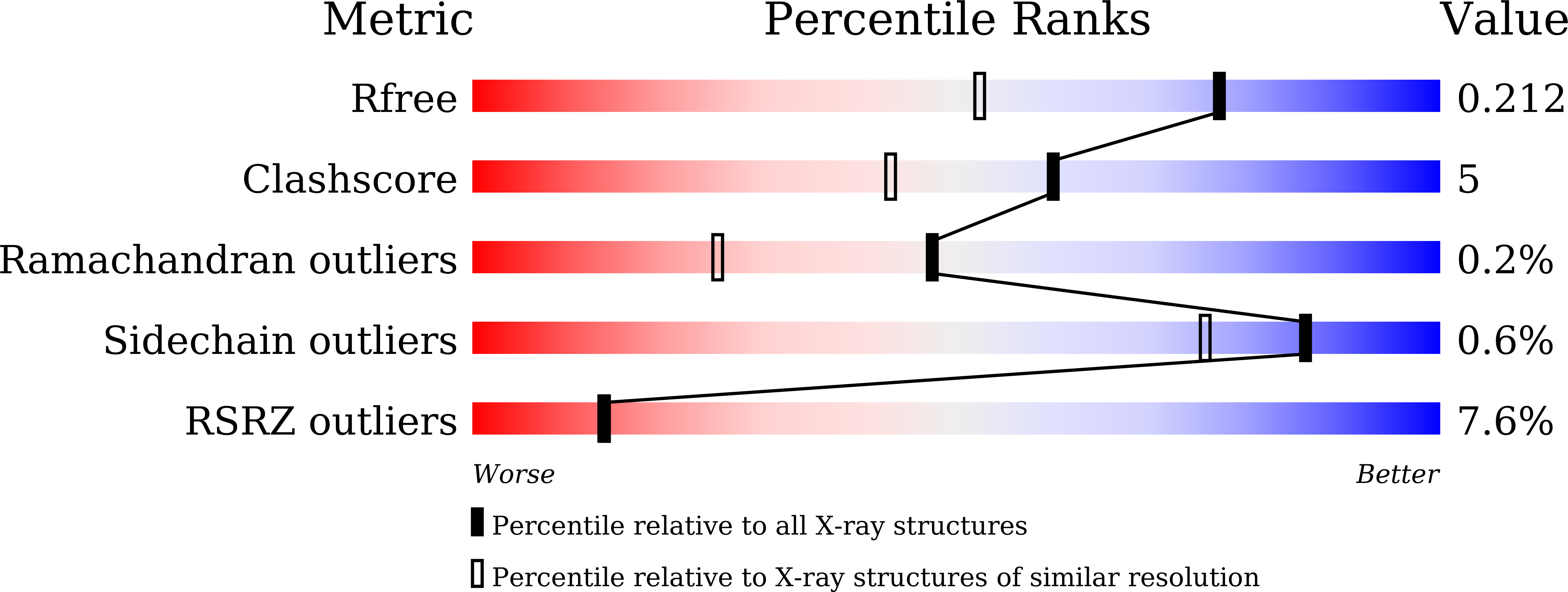
6M2O, 6M2U - PubMed Abstract:
Plant type III polyketide synthases produce diverse bioactive molecules with a great medicinal significance to human diseases. Here, we demonstrated versatility of a stilbene synthase (STS) from Pinus Sylvestris , which can accept various non-physiological substrates to form unnatural polyketide products. Three enzymes (4-coumarate CoA ligase, malonyl-CoA synthetase and engineered benzoate CoA ligase) along with synthetic chemistry was practiced to synthesize starter and extender substrates for STS. Of these, the crystal structures of benzoate CoA ligase (BadA) from Rhodopseudomonas palustris in an apo form or in complex with a 2-chloro-1,3-thiazole-5-carboxyl-AMP or 2-methylthiazole-5-carboxyl-AMP intermediate were determined at resolutions of 1.57 Å, 1.7 Å, and 2.13 Å, respectively, which reinforces its capacity in production of unusual CoA starters. STS exhibits broad substrate promiscuity effectively affording structurally diverse polyketide products. Seven novel products showed desired cytotoxicity against a panel of cancer cell lines (A549, HCT116, Cal27). With the treatment of two selected compounds, the cancer cells underwent cell apoptosis in a dose-dependent manner. The precursor-directed biosynthesis alongside structure-guided enzyme engineering greatly expands the pharmaceutical repertoire of lead compounds with promising/enhanced biological activities.
Organizational Affiliation:
Genomics Research Center, Academia Sinica, Taipei 11529, Taiwan.