Interception of teicoplanin oxidation intermediates yields new antimicrobial scaffolds.
Liu, Y.C., Li, Y.S., Lyu, S.Y., Hsu, L.J., Chen, Y.H., Huang, Y.T., Chan, H.C., Huang, C.J., Chen, G.H., Chou, C.C., Tsai, M.D., Li, T.L.(2011) Nat Chem Biol 7: 304-309
- PubMed: 21478878
- DOI: https://doi.org/10.1038/nchembio.556
- Primary Citation of Related Structures:
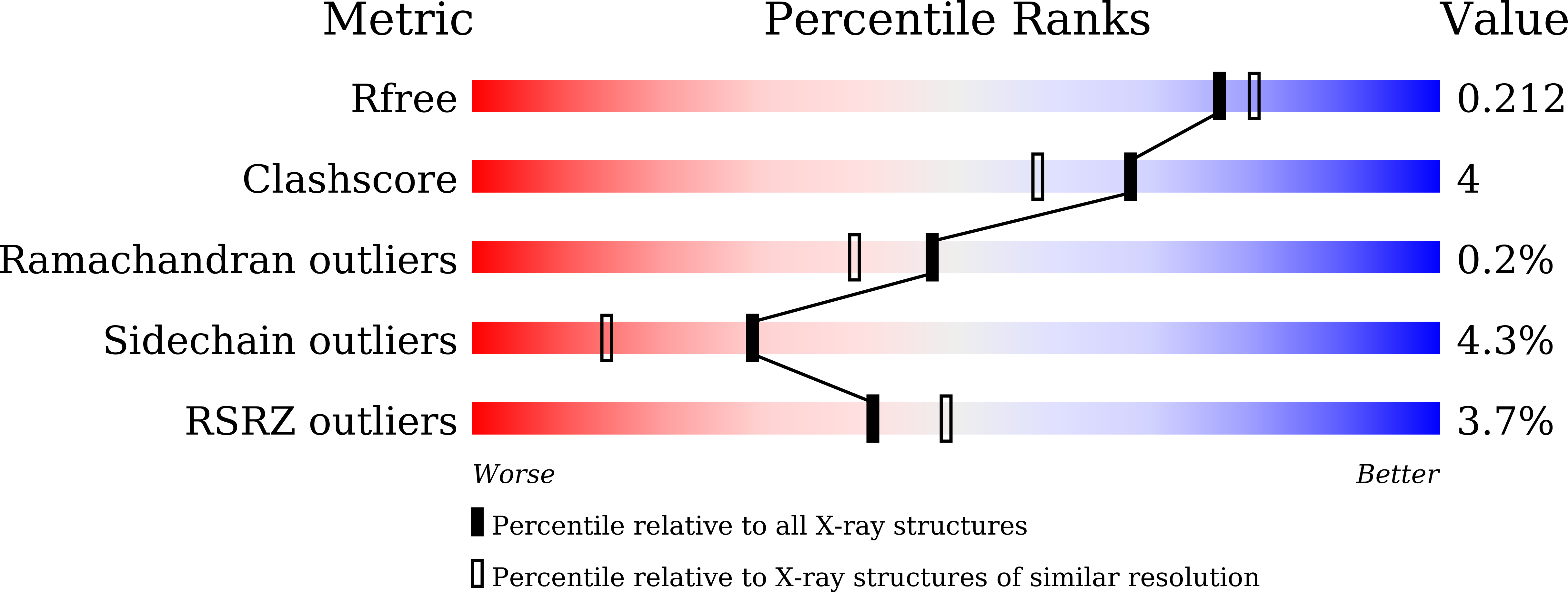
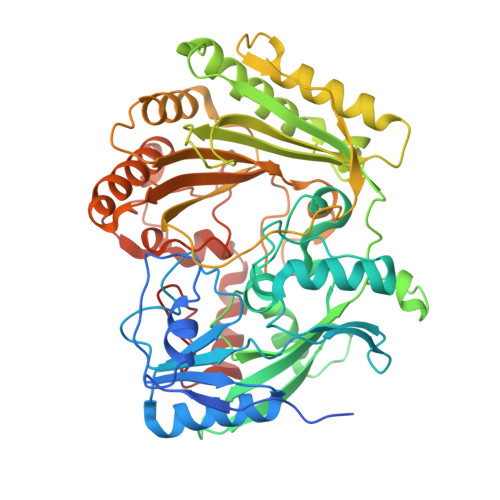
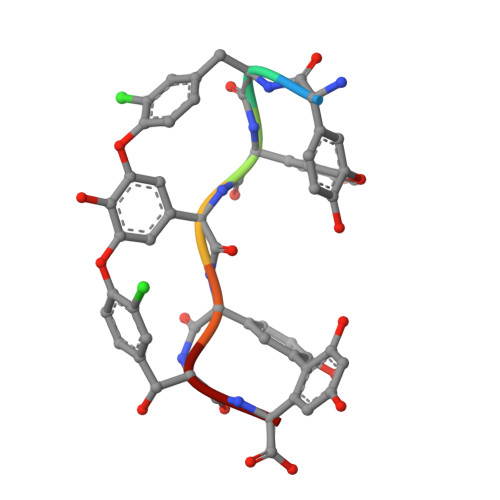
2WDW, 5AWV - PubMed Abstract:
In the search for new efficacious antibiotics, biosynthetic engineering offers attractive opportunities to introduce minor alterations to antibiotic structures that may overcome resistance. Dbv29, a flavin-containing oxidase, catalyzes the four-electron oxidation of a vancomycin-like glycopeptide to yield A40926. Structural and biochemical examination of Dbv29 now provides insights into residues that govern flavinylation and activity, protein conformation and reaction mechanism. In particular, the serendipitous discovery of a reaction intermediate in the crystal structure led us to identify an unexpected opportunity to intercept the normal enzyme mechanism at two different points to create new teicoplanin analogs. Using this method, we synthesized families of antibiotic analogs with amidated and aminated lipid chains, some of which showed marked potency and efficacy against multidrug resistant pathogens. This method offers a new strategy for the development of chemical diversity to combat antibacterial resistance.
Organizational Affiliation:
Genomics Research Center, Academia Sinica, Taipei, Taiwan.