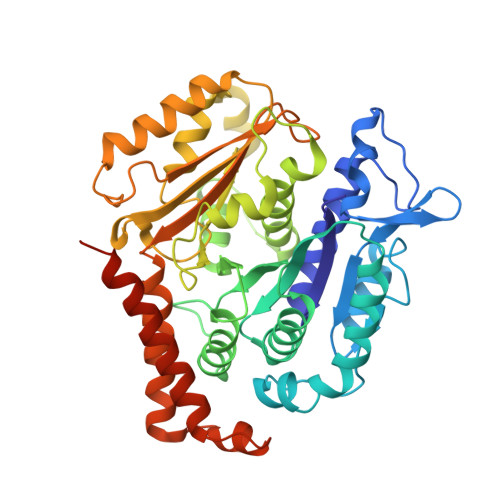
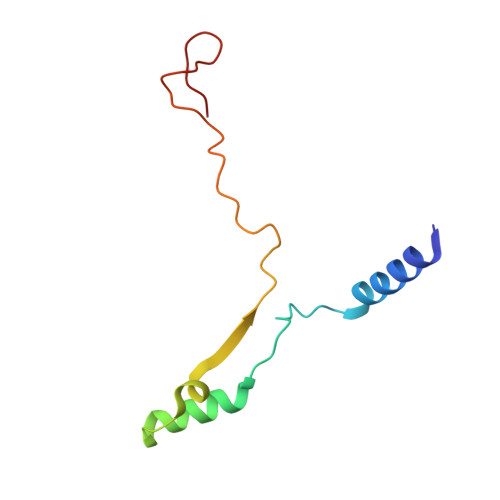
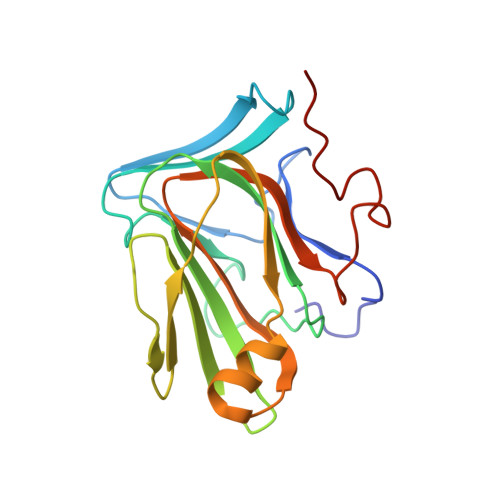
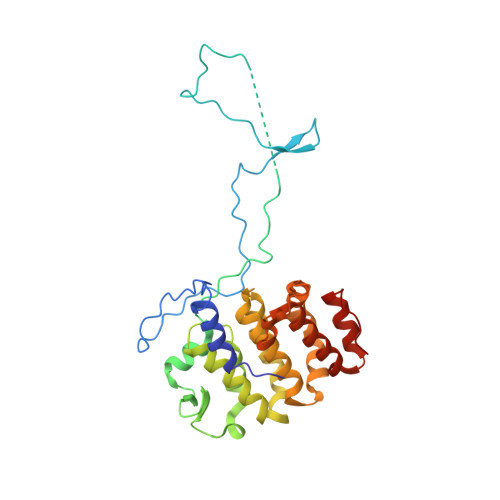
The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications.
Khalifa, A.A.Z., Ichikawa, M., Dai, D., Kubo, S., Black, C., Peri, K., McAlear, T.S., Veyron, S., Yang, S.K., Vargas, J., Bechstedt, S., Trempe, J.F., Bui, K.H.(2020) Elife 9
- PubMed: 31951202
- DOI: https://doi.org/10.7554/eLife.52760
- Primary Citation of Related Structures:
6VE7 - PubMed Abstract:
Microtubules are cytoskeletal structures involved in stability, transport and organization in the cell. The building blocks, the α- and β-tubulin heterodimers, form protofilaments that associate laterally into the hollow microtubule. Microtubule also exists as highly stable doublet microtubules in the cilia where stability is needed for ciliary beating and function. The doublet microtubule maintains its stability through interactions at its inner and outer junctions where its A- and B-tubules meet. Here, using cryo-electron microscopy, bioinformatics and mass spectrometry of the doublets of Chlamydomonas reinhardtii and Tetrahymena thermophila , we identified two new inner junction proteins, FAP276 and FAP106, and an inner junction-associated protein, FAP126, thus presenting the complete answer to the inner junction identity and localization. Our structural study of the doublets shows that the inner junction serves as an interaction hub that involves tubulin post-translational modifications. These interactions contribute to the stability of the doublet and hence, normal ciliary motility.
Organizational Affiliation:
Department of Anatomy and Cell Biology, McGill University, Québec, Canada.