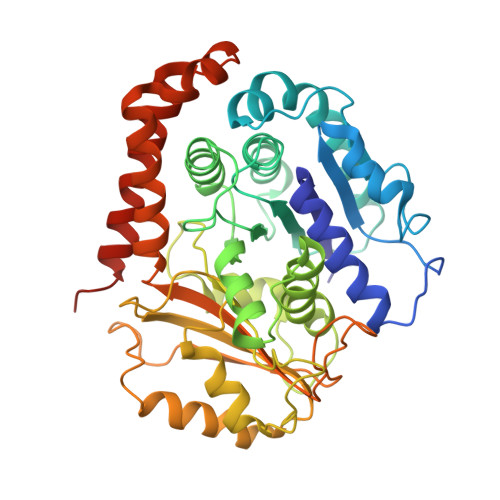
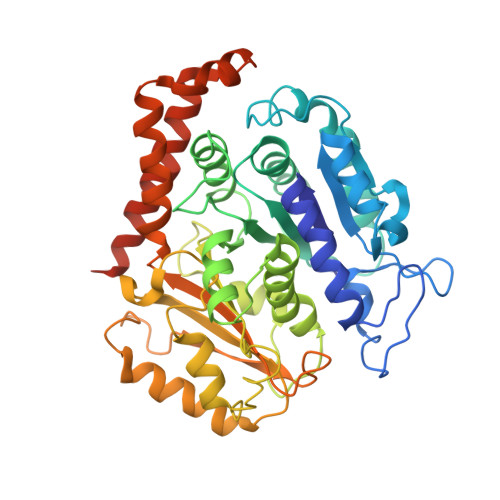
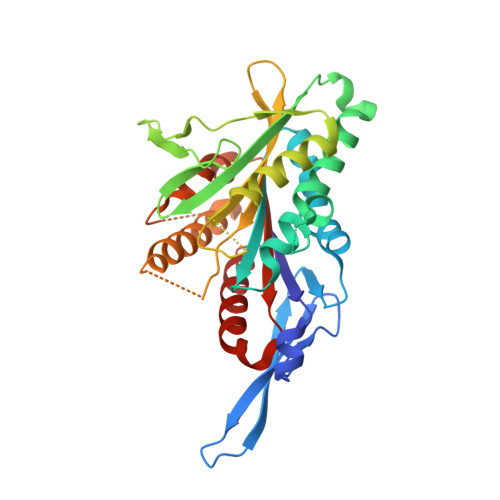
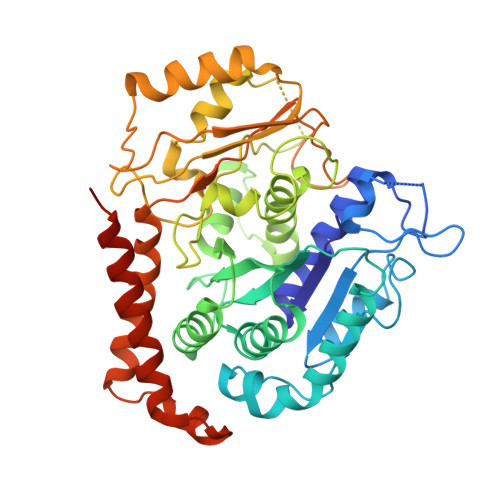
Structure of the kinesin13-microtubule ring complex.
Tan, D., Rice, W.J., Sosa, H.(2008) Structure 16: 1732-1739
- PubMed: 19000825
- DOI: https://doi.org/10.1016/j.str.2008.08.017
- Primary Citation of Related Structures:
3EDL - PubMed Abstract:
To investigate the mechanism of kinesin13-induced microtubule depolymerization, we have calculated a three-dimensional (3D) map of the kinesin13-microtubule ring complex, using cryo-electron microscopy (cryo-EM) and image analysis. An atomic model of the complex was produced by docking the crystal structures of tubulin and a kinesin13 motor domain (MD) into the 3D map. The model reveals a snapshot of the depolymerization mechanism by providing a 3D view of the complex formed between the kinesin13 MD and a curved tubulin protofilament (pf). It suggests that contacts mediated by kinesin13 class-specific residues in the putative microtubule-binding site stabilize intra-dimer tubulin curvature. In addition, a tubulin-binding site on the kinesin13 MD was identified. Mutations at this class-conserved site selectively disrupt the formation of microtubule-associated ring complexes.
- Department of Physiology and Biophysics, Albert Einstein College of Medicine, Bronx NY, 10461, USA.
Organizational Affiliation: