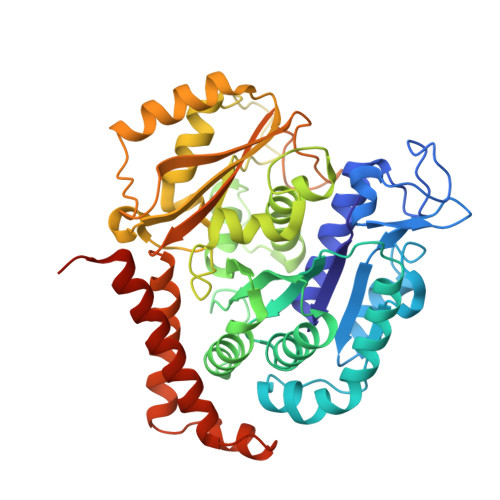
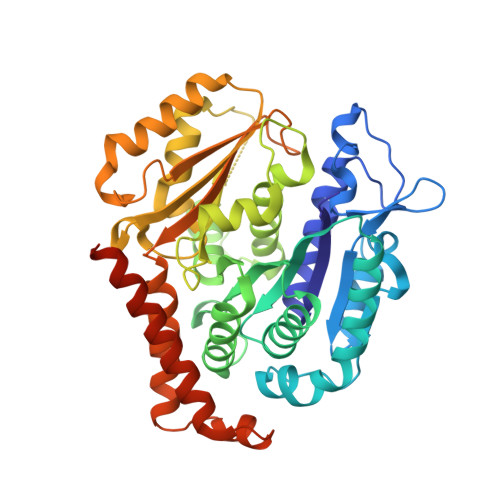
The Novel Microtubule-Destabilizing Drug BAL27862 Binds to the Colchicine Site of Tubulin with Distinct Effects on Microtubule Organization.
Prota, A.E., Danel, F., Bachmann, F., Bargsten, K., Buey, R.M., Pohlmann, J., Reinelt, S., Lane, H., Steinmetz, M.O.(2014) J Mol Biology 426: 1848-1860
- PubMed: 24530796
- DOI: https://doi.org/10.1016/j.jmb.2014.02.005
- Primary Citation of Related Structures:
4O2A, 4O2B - PubMed Abstract:
Microtubule-targeting agents are widely used for the treatment of cancer and as tool compounds to study the microtubule cytoskeleton. BAL27862 is a novel microtubule-destabilizing drug that is currently undergoing phase I clinical evaluation as the prodrug BAL101553. The drug is a potent inhibitor of tumor cell growth and shows a promising activity profile in a panel of human cancer models resistant to clinically relevant microtubule-targeting agents. Here, we evaluated the molecular mechanism of the tubulin-BAL27862 interaction using a combination of cell biology, biochemistry and structural biology methods. Tubulin-binding assays revealed that BAL27862 potently inhibited tubulin assembly at 37 °C with an IC50 of 1.4 μM and bound to unassembled tubulin with a stoichiometry of 1 mol/mol tubulin and a dissociation constant of 244±30 nM. BAL27862 bound to tubulin independently of vinblastine, without the formation of tubulin oligomers. The kinetics of BAL27862 binding to tubulin were distinct from those of colchicine, with evidence of competition between BAL27862 and colchicine for binding. Determination of the tubulin-BAL27862 structure by X-ray crystallography demonstrated that BAL27862 binds to the same site as colchicine at the intradimer interface. Comparison of crystal structures of tubulin-BAL27862 and tubulin-colchicine complexes shows that the binding mode of BAL27862 to tubulin is similar to that of colchicine. However, comparative analyses of the effects of BAL27862 and colchicine on the microtubule mitotic spindle and in tubulin protease-protection experiments suggest different outcomes of tubulin binding. Taken together, our data define BAL27862 as a potent, colchicine site-binding, microtubule-destabilizing agent with distinct effects on microtubule organization.
- Laboratory of Biomolecular Research, Department of Biology and Chemistry Paul Scherrer Institut, 5232 Villigen, Switzerland.
Organizational Affiliation: