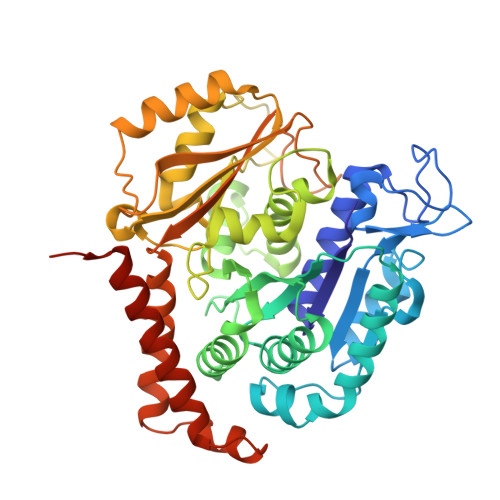
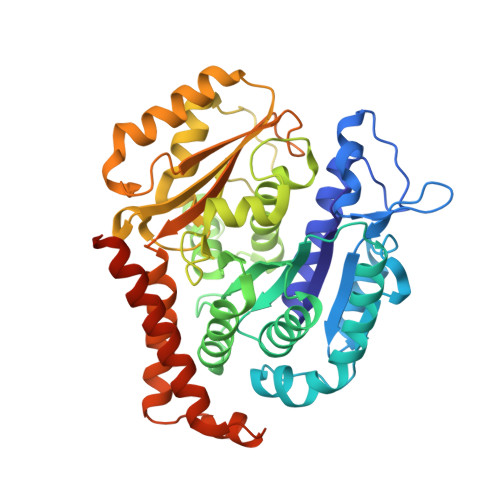
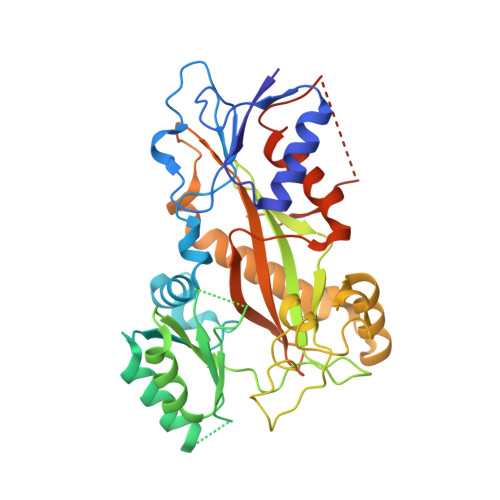
Termination of Protofilament Elongation by Eribulin Induces Lattice Defects that Promote Microtubule Catastrophes.
Doodhi, H., Prota, A.E., Rodriguez-Garcia, R., Xiao, H., Custar, D.W., Bargsten, K., Katrukha, E.A., Hilbert, M., Hua, S., Jiang, K., Grigoriev, I., Yang, C.P., Cox, D., Horwitz, S.B., Kapitein, L.C., Akhmanova, A., Steinmetz, M.O.(2016) Curr Biol 26: 1713-1721
- PubMed: 27321995
- DOI: https://doi.org/10.1016/j.cub.2016.04.053
- Primary Citation of Related Structures:
5JH7 - PubMed Abstract:
Microtubules are dynamic polymers built of tubulin dimers that attach in a head-to-tail fashion to form protofilaments, which further associate laterally to form a tube. Asynchronous elongation of individual protofilaments can potentially lead to an altered microtubule-end structure that promotes sudden depolymerization, termed catastrophe [1-4]. However, how the dynamics of individual protofilaments relates to overall growth persistence has remained unclear. Here, we used the microtubule targeting anti-cancer drug Eribulin [5-7] to explore the consequences of stalled protofilament elongation on microtubule growth. Using X-ray crystallography, we first revealed that Eribulin binds to a site on β-tubulin that is required for protofilament plus-end elongation. Based on the structural information, we engineered a fluorescent Eribulin molecule. We demonstrate that single Eribulin molecules specifically interact with microtubule plus ends and are sufficient to either trigger a catastrophe or induce slow and erratic microtubule growth in the presence of EB3. Interestingly, we found that Eribulin increases the frequency of EB3 comet "splitting," transient events where a slow and erratically progressing comet is followed by a faster comet. This observation possibly reflects the "healing" of a microtubule lattice. Because EB3 comet splitting was also observed in control microtubules in the absence of any drugs, we propose that Eribulin amplifies a natural pathway toward catastrophe by promoting the arrest of protofilament elongation.
Organizational Affiliation:
Cell Biology, Department of Biology, Faculty of Science, Utrecht University, 3584 CH Utrecht, the Netherlands.