Synthesis, biological evaluation and X-ray structure of anti-microtubule agents
Chu, Y., Wang, Y., Yang, J., Li, W.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin alpha chain | 450 | Sus barbatus | Mutation(s): 0 | 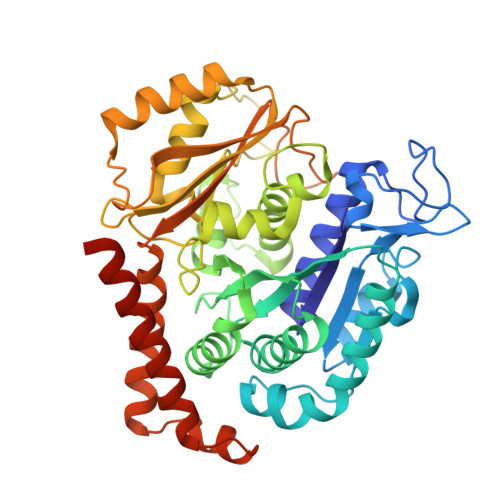 | |
UniProt | |||||
Find proteins for A0A0R4I993 (Sus barbatus) Explore A0A0R4I993 Go to UniProtKB: A0A0R4I993 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0R4I993 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin beta chain | 445 | Sus barbatus | Mutation(s): 0 | 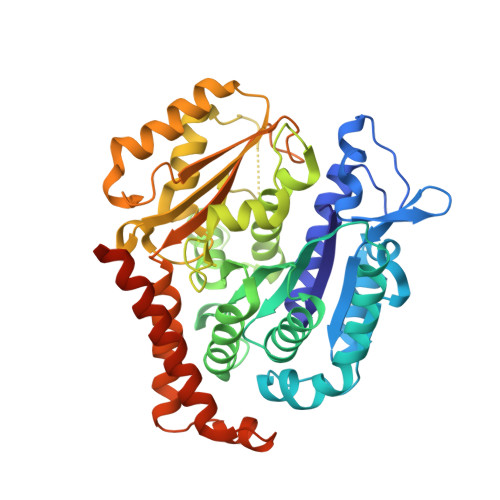 | |
UniProt | |||||
Find proteins for A0A0R4I995 (Sus barbatus) Explore A0A0R4I995 Go to UniProtKB: A0A0R4I995 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0R4I995 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Stathmin-4 | 184 | Rattus norvegicus | Mutation(s): 0 Gene Names: Stmn4 | 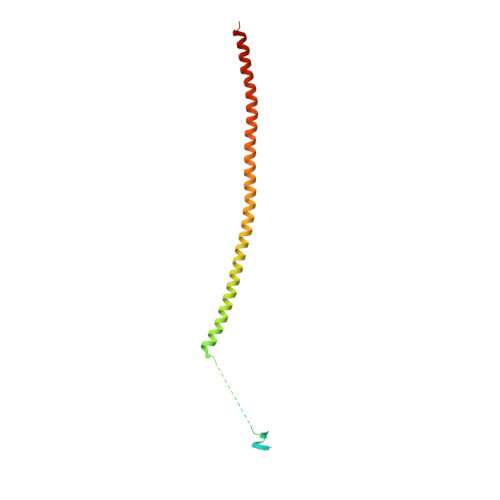 | |
UniProt | |||||
Find proteins for P63043 (Rattus norvegicus) Explore P63043 Go to UniProtKB: P63043 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63043 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TUBULIN TYROSINE LIGASE | 384 | Gallus gallus | Mutation(s): 0 Gene Names: TTL | 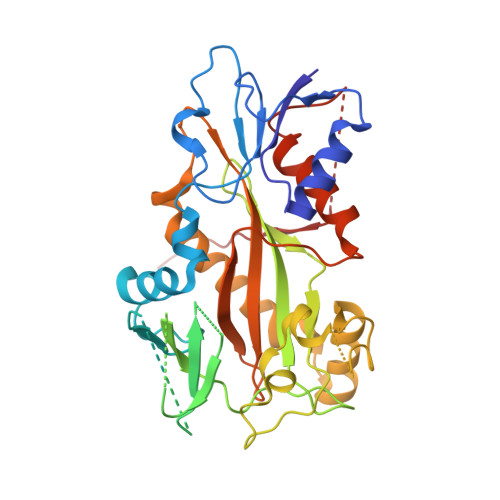 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GTP Query on GTP | G [auth A], P [auth C], S [auth D] | GUANOSINE-5'-TRIPHOSPHATE C10 H16 N5 O14 P3 XKMLYUALXHKNFT-UUOKFMHZSA-N |  | ||
| ACP Query on ACP | U [auth F] | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER C11 H18 N5 O12 P3 UFZTZBNSLXELAL-IOSLPCCCSA-N |  | ||
| 87U Query on 87U | M [auth B] | (3Z,6Z)-3-[(4-tert-butyl-1H-imidazol-5-yl)methylidene]-6-[[3-(4-fluorophenyl)carbonylphenyl]methylidene]piperazine-2,5-dione C26 H23 F N4 O3 PPBYGOIXJTYVGU-FDYZEBBJSA-N |  | ||
| GDP Query on GDP | J [auth B] | GUANOSINE-5'-DIPHOSPHATE C10 H15 N5 O11 P2 QGWNDRXFNXRZMB-UUOKFMHZSA-N |  | ||
| MES Query on MES | L [auth B], O [auth B] | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID C6 H13 N O4 S SXGZJKUKBWWHRA-UHFFFAOYSA-N |  | ||
| CA Query on CA | H [auth A], N [auth B], R [auth C] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| MG Query on MG | I [auth B], K [auth B], Q [auth C], T [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 105.257 | α = 90 |
| b = 157.885 | β = 90 |
| c = 182.427 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| SCALEPACK | data reduction |
| DM | model building |
| SHARP | model building |
| DENZO | data collection |
| SHELXD | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Qingdao science and technology bureau | China | 16-5-1-61-jch |