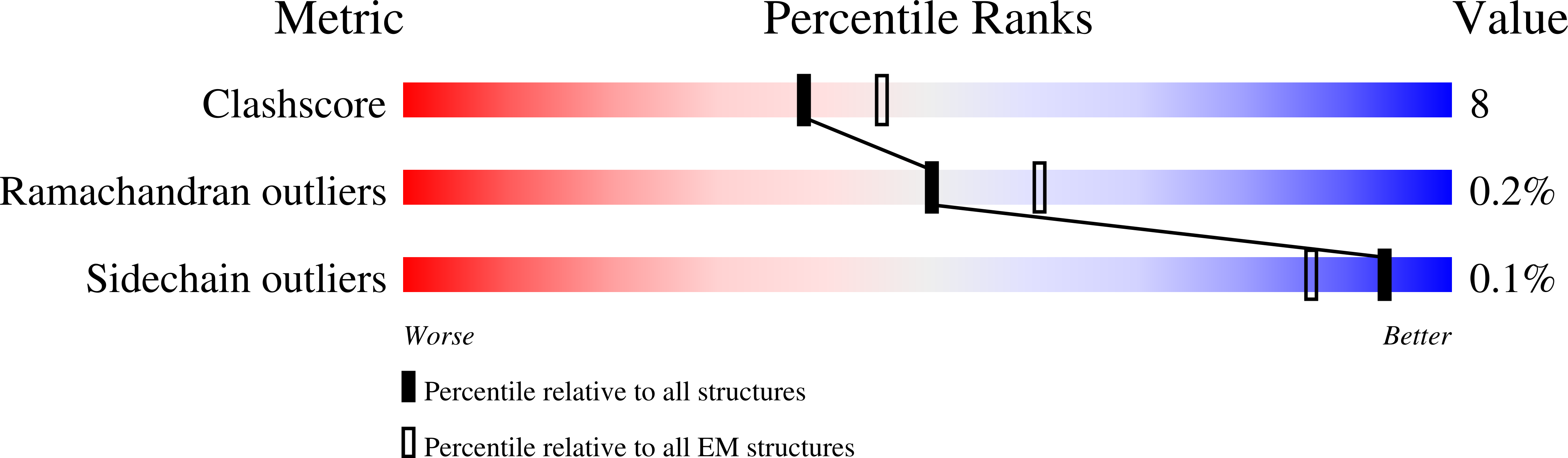Transposon molecular domestication and the evolution of the RAG recombinase.
Zhang, Y., Cheng, T.C., Huang, G., Lu, Q., Surleac, M.D., Mandell, J.D., Pontarotti, P., Petrescu, A.J., Xu, A., Xiong, Y., Schatz, D.G.(2019) Nature 569: 79-84
- PubMed: 30971819
- DOI: https://doi.org/10.1038/s41586-019-1093-7
- Primary Citation of Related Structures:
6B40 - PubMed Abstract:
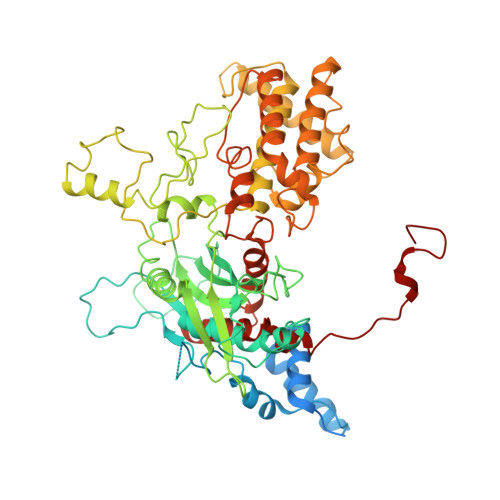
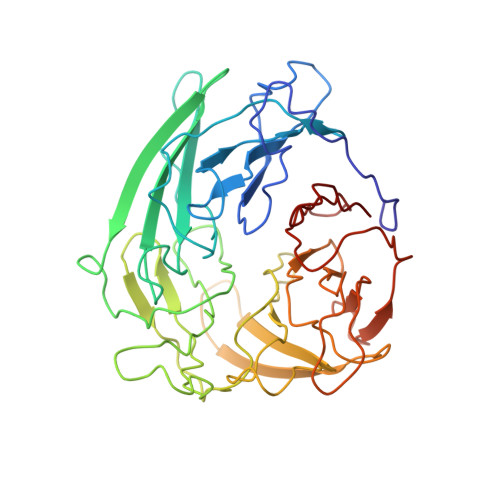
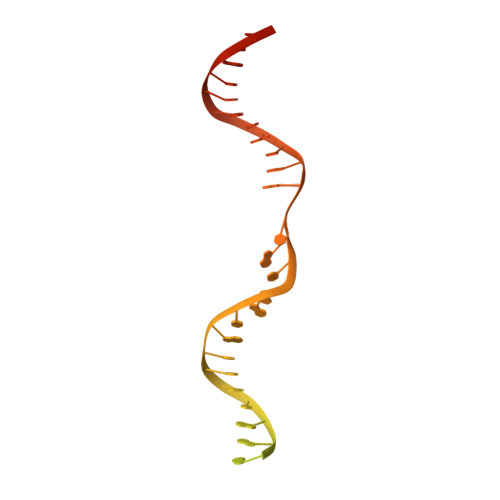
Domestication of a transposon (a DNA sequence that can change its position in a genome) to give rise to the RAG1-RAG2 recombinase (RAG) and V(D)J recombination, which produces the diverse repertoire of antibodies and T cell receptors, was a pivotal event in the evolution of the adaptive immune system of jawed vertebrates. The evolutionary adaptations that transformed the ancestral RAG transposase into a RAG recombinase with appropriately regulated DNA cleavage and transposition activities are not understood. Here, beginning with cryo-electron microscopy structures of the amphioxus ProtoRAG transposase (an evolutionary relative of RAG), we identify amino acid residues and domains the acquisition or loss of which underpins the propensity of RAG for coupled cleavage, its preference for asymmetric DNA substrates and its inability to perform transposition in cells. In particular, we identify two adaptations specific to jawed-vertebrates-arginine 848 in RAG1 and an acidic region in RAG2-that together suppress RAG-mediated transposition more than 1,000-fold. Our findings reveal a two-tiered mechanism for the suppression of RAG-mediated transposition, illuminate the evolution of V(D)J recombination and provide insight into the principles that govern the molecular domestication of transposons.
Organizational Affiliation:
Department of Immunobiology, Yale School of Medicine, New Haven, CT, USA.