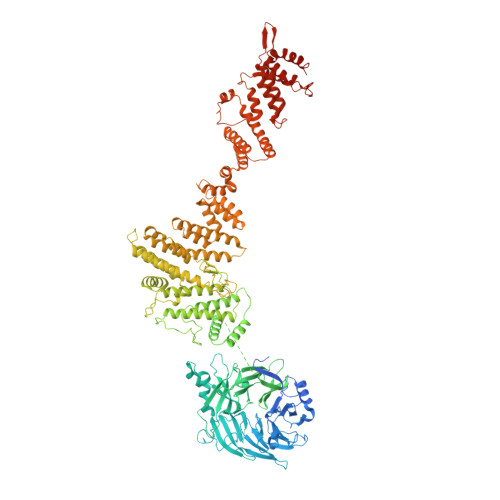
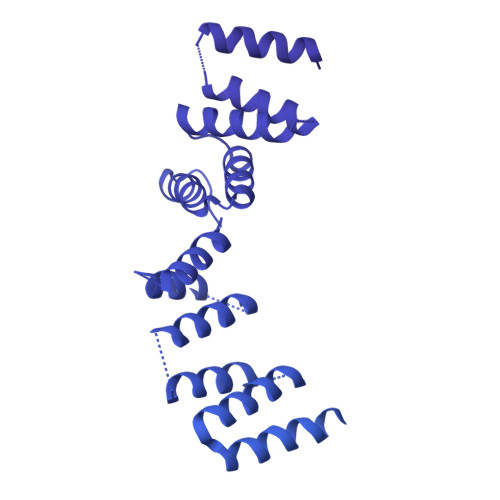
Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis.
Huang, G., Zhang, Y., Zhu, X., Zeng, C., Wang, Q., Zhou, Q., Tao, Q., Liu, M., Lei, J., Yan, C., Shi, Y.(2020) Cell Res 30: 520-531
- PubMed: 32376910
- DOI: https://doi.org/10.1038/s41422-020-0319-4
- Primary Citation of Related Structures:
6LK8 - PubMed Abstract:
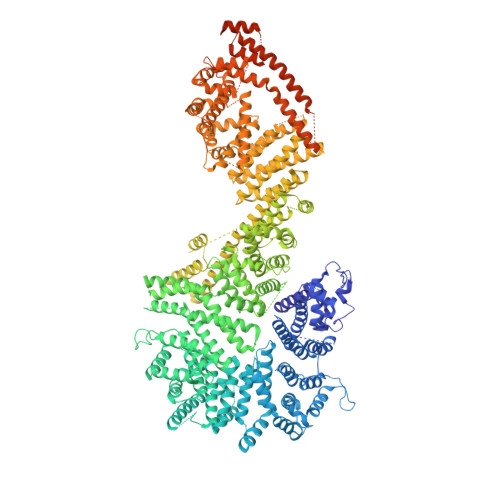
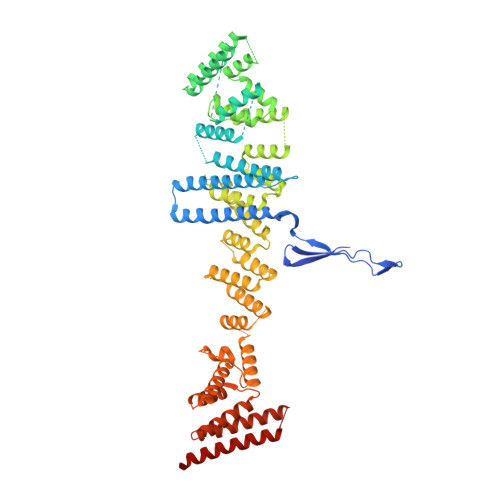
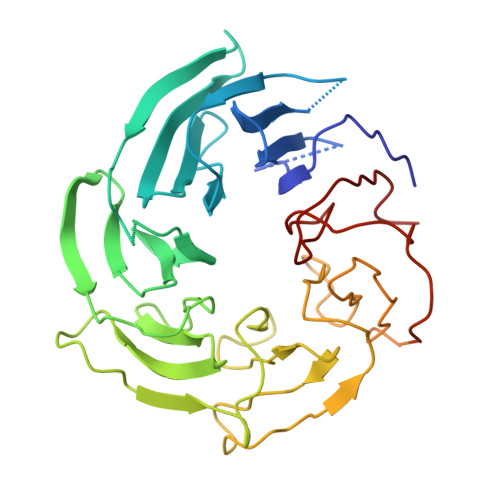
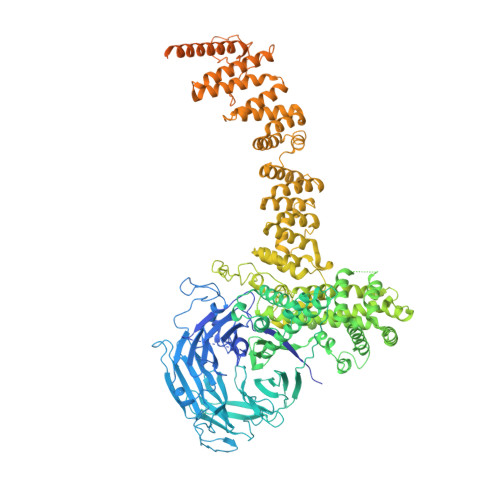
The nuclear pore complex (NPC) exhibits structural plasticity and has only been characterized at local resolutions of up to 15 Å for the cytoplasmic ring (CR). Here we present a single-particle cryo-electron microscopy (cryo-EM) structure of the CR from Xenopus laevis NPC at average resolutions of 5.5-7.9 Å, with local resolutions reaching 4.5 Å. Improved resolutions allow identification and placement of secondary structural elements in the majority of the CR components. The two Y complexes in each CR subunit interact with each other and associate with those from flanking subunits, forming a circular scaffold. Within each CR subunit, the Nup358-containing region wraps around the stems of both Y complexes, likely stabilizing the scaffold. Nup205 connects the short arms of the two Y complexes and associates with the stem of a neighboring Y complex. The Nup214-containing region uses an extended coiled-coil to link Nup85 of the two Y complexes and protrudes into the axial pore of the NPC. These previously uncharacterized structural features reveal insights into NPC assembly.
- Beijing Advanced Innovation Center for Structural Biology & Frontier Research Center for Biological Structure, School of Life Sciences, Tsinghua University, Beijing, 100084, China.
Organizational Affiliation: