A Ubiquitin-Binding Domain that Binds a Structural Fold Distinct from that of Ubiquitin.
Lim, M., Newman, J.A., Williams, H.L., Masino, L., Aitkenhead, H., Gravard, A.E., Gileadi, O., Svejstrup, J.Q.(2019) Structure
- PubMed: 31204252
- DOI: https://doi.org/10.1016/j.str.2019.05.003
- Primary Citation of Related Structures:
6H3A, 6QU1 - PubMed Abstract:
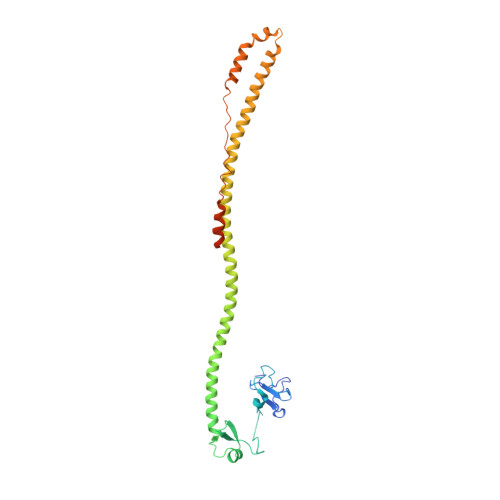
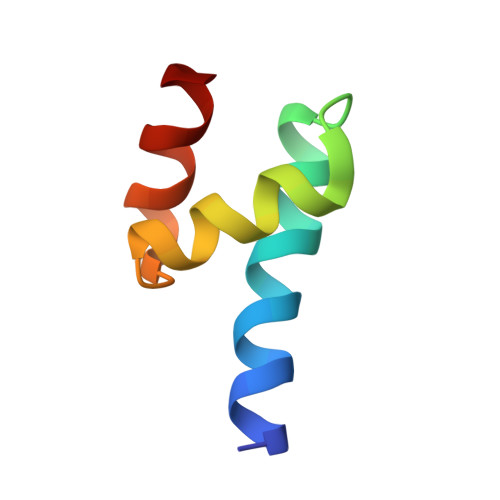
Ubiquitylation, the posttranslational linkage of ubiquitin moieties to lysines in target proteins, helps regulate a myriad of biological processes. Ubiquitin, and sometimes ubiquitin-homology domains, are recognized by ubiquitin-binding domains, including CUE domains. CUE domains are thus generally thought to function by mediating interactions with ubiquitylated proteins. The chromatin remodeler, SMARCAD1, interacts with KAP1, a transcriptional corepressor. The SMARCAD1-KAP1 interaction is direct and involves the first SMARCAD1 CUE domain (CUE1) and the RBCC domain of KAP1. Here, we present a structural model of the KAP1 RBCC-SMARCAD1 CUE1 complex based on X-ray crystallography. Remarkably, CUE1, a canonical CUE domain, recognizes a cluster of exposed hydrophobic and surrounding charged/amphipathic residues on KAP1, which are presented in the context of a coiled-coil domain, not in a structure resembling ubiquitin. Together, these data suggest that CUE domains may have a wider function than simply recognizing ubiquitin and the ubiquitin-fold.
Organizational Affiliation:
Mechanisms of Transcription Laboratory, The Francis Crick Institute, 1 Midland Road, London NW1 1AT, UK.