Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Atherton, J., Luo, Y., Xiang, S., Yang, C., Rai, A., Jiang, K., Stangier, M., Vemu, A., Cook, A.D., Wang, S., Roll-Mecak, A., Steinmetz, M.O., Akhmanova, A., Baldus, M., Moores, C.A.(2019) Nat Commun 10: 5236-5236
- PubMed: 31748546
- DOI: https://doi.org/10.1038/s41467-019-13247-6
- Primary Citation of Related Structures:
6QUS, 6QUY, 6QVE, 6QVJ - PubMed Abstract:
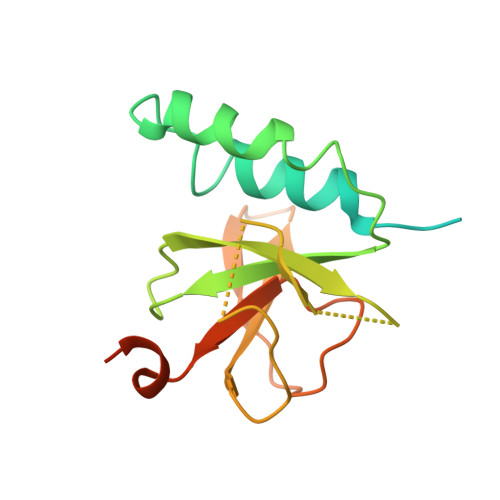
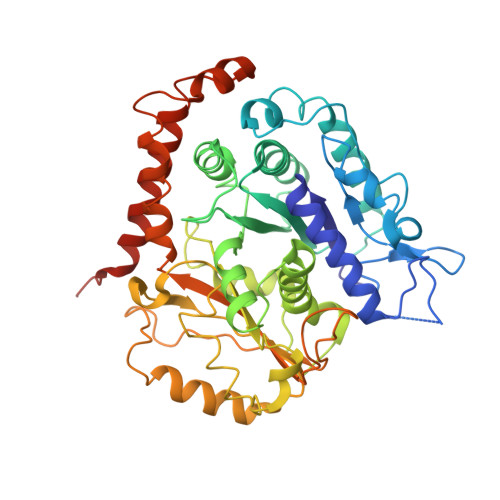
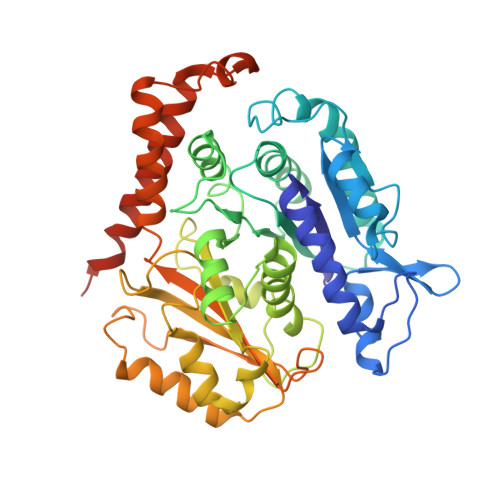
CAMSAP/Patronins regulate microtubule minus-end dynamics. Their end specificity is mediated by their CKK domains, which we proposed recognise specific tubulin conformations found at minus ends. To critically test this idea, we compared the human CAMSAP1 CKK domain (HsCKK) with a CKK domain from Naegleria gruberi (NgCKK), which lacks minus-end specificity. Here we report near-atomic cryo-electron microscopy structures of HsCKK- and NgCKK-microtubule complexes, which show that these CKK domains share the same protein fold, bind at the intradimer interprotofilament tubulin junction, but exhibit different footprints on microtubules. NMR experiments show that both HsCKK and NgCKK are remarkably rigid. However, whereas NgCKK binding does not alter the microtubule architecture, HsCKK remodels its microtubule interaction site and changes the underlying polymer structure because the tubulin lattice conformation is not optimal for its binding. Thus, in contrast to many MAPs, the HsCKK domain can differentiate subtly specific tubulin conformations to enable microtubule minus-end recognition.
Organizational Affiliation:
Institute of Structural and Molecular Biology, Birkbeck, University of London, Malet Street, London, UK. j.atherton@mail.cryst.bbk.ac.uk.