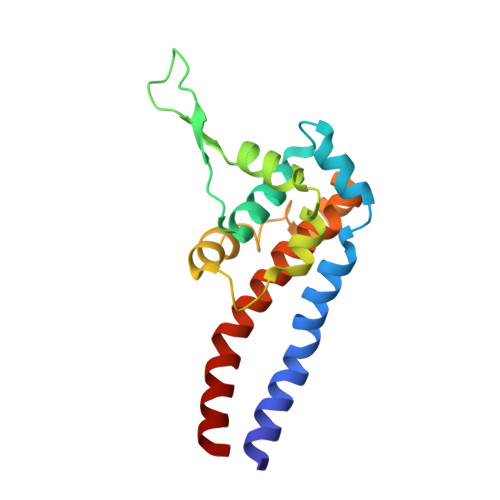
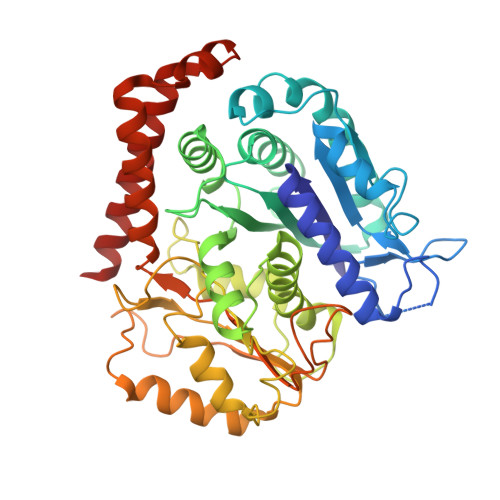
Cryo-EM of dynein microtubule-binding domains shows how an axonemal dynein distorts the microtubule.
Lacey, S.E., He, S., Scheres, S.H., Carter, A.P.(2019) Elife 8
- PubMed: 31264960
- DOI: https://doi.org/10.7554/eLife.47145
- Primary Citation of Related Structures:
6RZA, 6RZB - PubMed Abstract:
Dyneins are motor proteins responsible for transport in the cytoplasm and the beating of axonemes in cilia and flagella. They bind and release microtubules via a compact microtubule-binding domain (MTBD) at the end of a coiled-coil stalk. We address how cytoplasmic and axonemal dynein MTBDs bind microtubules at near atomic resolution. We decorated microtubules with MTBDs of cytoplasmic dynein-1 and axonemal dynein DNAH7 and determined their cryo-EM structures using helical Relion. The majority of the MTBD is rigid upon binding, with the transition to the high-affinity state controlled by the movement of a single helix at the MTBD interface. DNAH7 contains an 18-residue insertion, found in many axonemal dyneins, that contacts the adjacent protofilament. Unexpectedly, we observe that DNAH7, but not dynein-1, induces large distortions in the microtubule cross-sectional curvature. This raises the possibility that dynein coordination in axonemes is mediated via conformational changes in the microtubule.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge, United Kingdom.