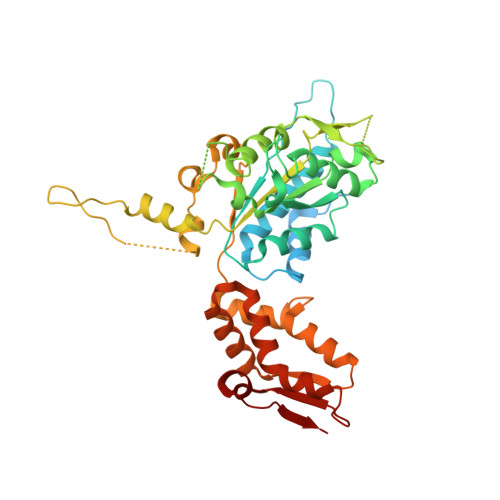
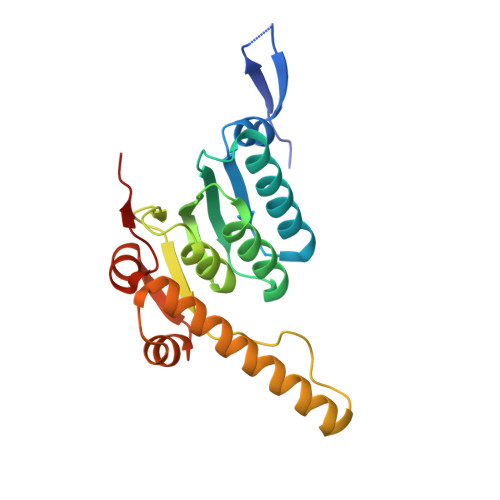
A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Ripstein, Z.A., Vahidi, S., Houry, W.A., Rubinstein, J.L., Kay, L.E.(2020) Elife 9
- PubMed: 31916936
- DOI: https://doi.org/10.7554/eLife.52158
- Primary Citation of Related Structures:
6VFS, 6VFX - PubMed Abstract:
The ClpXP degradation machine consists of a hexameric AAA+ unfoldase (ClpX) and a pair of heptameric serine protease rings (ClpP) that unfold, translocate, and subsequently degrade client proteins. ClpXP is an important target for drug development against infectious diseases. Although structures are available for isolated ClpX and ClpP rings, it remains unknown how symmetry mismatched ClpX and ClpP work in tandem for processive substrate translocation into the ClpP proteolytic chamber. Here, we present cryo-EM structures of the substrate-bound ClpXP complex from Neisseria meningitidis at 2.3 to 3.3 Å resolution. The structures allow development of a model in which the sequential hydrolysis of ATP is coupled to motions of ClpX loops that lead to directional substrate translocation and ClpX rotation relative to ClpP. Our data add to the growing body of evidence that AAA+ molecular machines generate translocating forces by a common mechanism.
- Department of Biochemistry, University of Toronto, Toronto, Canada.
Organizational Affiliation: