Dual role of a (p)ppGpp- and (p)ppApp-degrading enzyme in biofilm formation and interbacterial antagonism.
Steinchen, W., Ahmad, S., Valentini, M., Eilers, K., Majkini, M., Altegoer, F., Lechner, M., Filloux, A., Whitney, J.C., Bange, G.(2021) Mol Microbiol 115: 1339-1356
- PubMed: 33448498
- DOI: https://doi.org/10.1111/mmi.14684
- Primary Citation of Related Structures:
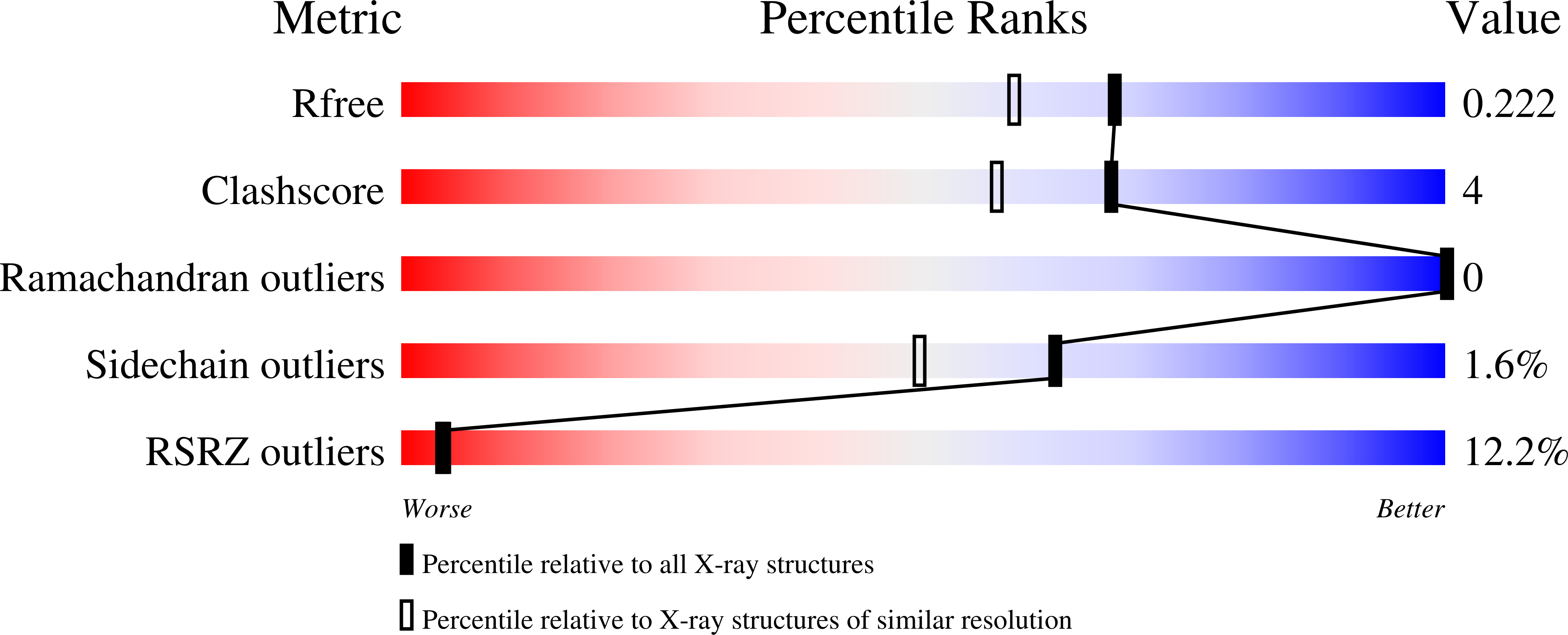
6YVC - PubMed Abstract:
The guanosine nucleotide-based second messengers ppGpp and pppGpp (collectively: (p)ppGpp) enable adaptation of microorganisms to environmental changes and stress conditions. In contrast, the closely related adenosine nucleotides (p)ppApp are involved in type VI secretion system (T6SS)-mediated killing during bacterial competition. Long RelA-SpoT Homolog (RSH) enzymes regulate synthesis and degradation of (p)ppGpp (and potentially also (p)ppApp) through their synthetase and hydrolase domains, respectively. Small alarmone hydrolases (SAH) that consist of only a hydrolase domain are found in a variety of bacterial species, including the opportunistic human pathogen Pseudomonas aeruginosa. Here, we present the structure and mechanism of P. aeruginosa SAH showing that the enzyme promiscuously hydrolyses (p)ppGpp and (p)ppApp in a strictly manganese-dependent manner. While being dispensable for P. aeruginosa growth or swimming, swarming, and twitching motilities, its enzymatic activity is required for biofilm formation. Moreover, (p)ppApp-degradation by SAH provides protection against the T6SS (p)ppApp synthetase effector Tas1, suggesting that SAH enzymes can also serve as defense proteins during interbacterial competition.
Organizational Affiliation:
Center for Synthetic Microbiology (SYNMIKRO) & Faculty of Chemistry, Philipps-University Marburg, Marburg, Germany.