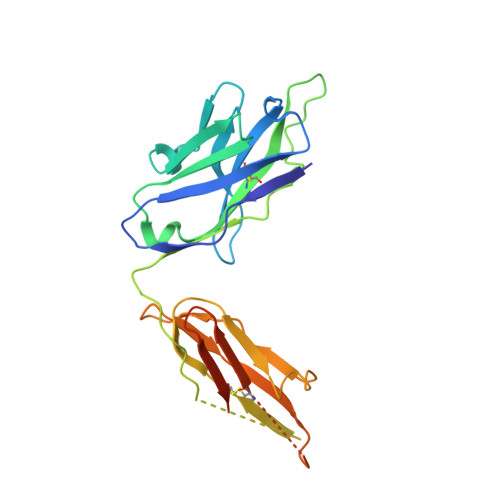
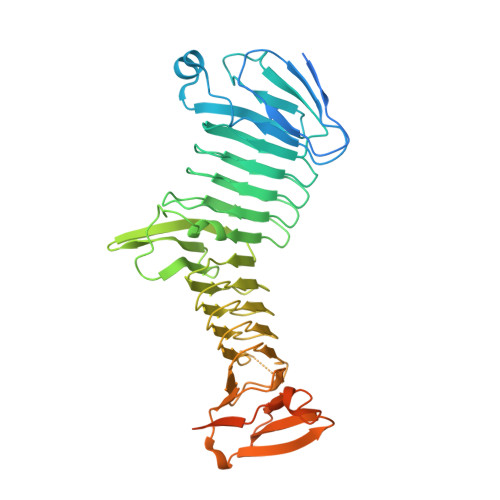
Structural basis for antibody binding to adenylate cyclase toxin reveals RTX linkers as neutralization-sensitive epitopes.
Goldsmith, J.A., DiVenere, A.M., Maynard, J.A., McLellan, J.S.(2021) PLoS Pathog 17: e1009920-e1009920
- PubMed: 34547035
- DOI: https://doi.org/10.1371/journal.ppat.1009920
- Primary Citation of Related Structures:
7RAH - PubMed Abstract:
RTX leukotoxins are a diverse family of prokaryotic virulence factors that are secreted by the type 1 secretion system (T1SS) and target leukocytes to subvert host defenses. T1SS substrates all contain a C-terminal RTX domain that mediates recruitment to the T1SS and drives secretion via a Brownian ratchet mechanism. Neutralizing antibodies against the Bordetella pertussis adenylate cyclase toxin, an RTX leukotoxin essential for B. pertussis colonization, have been shown to target the RTX domain and prevent binding to the αMβ2 integrin receptor. Knowledge of the mechanisms by which antibodies bind and neutralize RTX leukotoxins is required to inform structure-based design of bacterial vaccines, however, no structural data are available for antibody binding to any T1SS substrate. Here, we determine the crystal structure of an engineered RTX domain fragment containing the αMβ2-binding site bound to two neutralizing antibodies. Notably, the receptor-blocking antibodies bind to the linker regions of RTX blocks I-III, suggesting they are key neutralization-sensitive sites within the RTX domain and are likely involved in binding the αMβ2 receptor. As the engineered RTX fragment contained these key epitopes, we assessed its immunogenicity in mice and showed that it elicits similar neutralizing antibody titers to the full RTX domain. The results from these studies will support the development of bacterial vaccines targeting RTX leukotoxins, as well as next-generation B. pertussis vaccines.
Organizational Affiliation:
Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas, United States of America.