Crystal structure of HLA B*18:01 in complex with EEIEITTHF, an 9-mer epitope from Influenza A
Murdolo, L.D., Maddumaage, J., Gras, S.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MHC class I antigen | 276 | Homo sapiens | Mutation(s): 0 Gene Names: HLA-B | 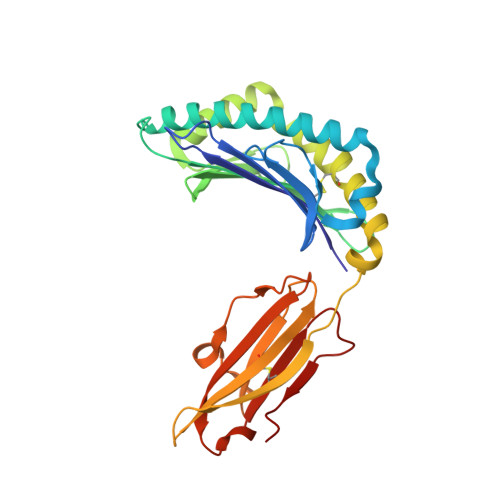 | |
UniProt | |||||
Find proteins for A0A167RQK8 (Homo sapiens) Explore A0A167RQK8 Go to UniProtKB: A0A167RQK8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A167RQK8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA-directed RNA polymerase catalytic subunit | B [auth C] | 9 | Influenza A virus (A/Memphis/18/1978(H3N2)) | Mutation(s): 0 EC: 2.7.7.48 | 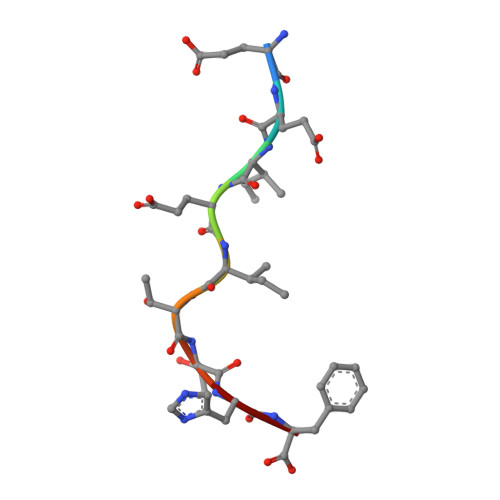 |
UniProt | |||||
Find proteins for Q2VNC5 (Influenza A virus (strain A/Memphis/18/1978 H3N2)) Explore Q2VNC5 Go to UniProtKB: Q2VNC5 | |||||
Entity Groups | |||||
| UniProt Group | Q2VNC5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-2-microglobulin | C [auth B] | 100 | Homo sapiens | Mutation(s): 0 Gene Names: B2M, CDABP0092, HDCMA22P | 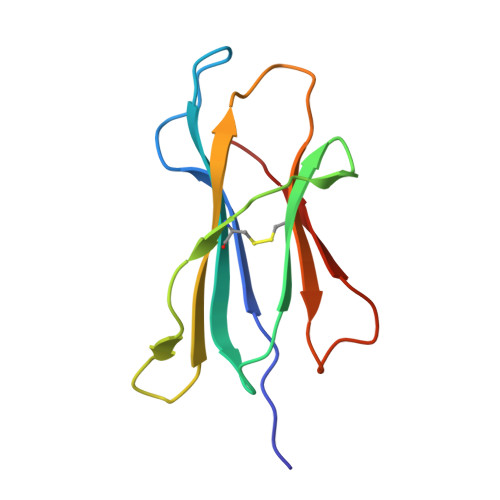 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61769 (Homo sapiens) Explore P61769 Go to UniProtKB: P61769 | |||||
PHAROS: P61769 GTEx: ENSG00000166710 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61769 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG (Subject of Investigation/LOI) Query on PEG | D [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| EDO (Subject of Investigation/LOI) Query on EDO | E [auth A], F [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 50.75 | α = 90 |
| b = 81.627 | β = 90 |
| c = 110.556 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Health and Medical Research Council (NHMRC, Australia) | Australia | -- |