Crystal structure of Lsd18 in complex with ligands
Deng, Y.M., Hu, Y.L., Chen, X.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Putative epoxidase LasC | 488 | Streptomyces lasalocidi | Mutation(s): 0 Gene Names: lsd18 EC: 1.14.13 | 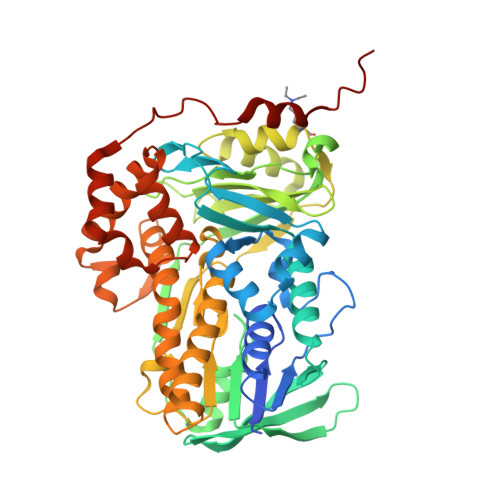 | |
UniProt | |||||
Find proteins for B5M9L6 (Streptomyces lasalocidi) Explore B5M9L6 Go to UniProtKB: B5M9L6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B5M9L6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD (Subject of Investigation/LOI) Query on FAD | C [auth A], H [auth B] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| X9E (Subject of Investigation/LOI) Query on X9E | F [auth A] | (4R,5S)-3-((2R,3S,4S,E)-2,6-diethyl-9-((2R,3R)-2-ethyl-3-methyloxiran-2-yl)-3-hydroxy-4-methylnon-6-enoyl)-4-methyl-5-phenyloxazolidin-2-one C29 H43 N O5 MAKHZPRKABYPIS-LKCQFNNRSA-N |  | ||
| A1LWM (Subject of Investigation/LOI) Query on A1LWM | K [auth B] | (4R,5S)-4-methyl-5-phenyl-3-((2R,3S,4S,6E,10E)-2,6,10-triethyl-3-hydroxy-4-methyldodeca-6,10-dienoyl)oxazolidin-2-one C29 H43 N O4 GIEHXLSOYWFSMM-FYYHNBHTSA-N |  | ||
| DMS (Subject of Investigation/LOI) Query on DMS | D [auth A], E [auth A], I [auth B], J [auth B] | DIMETHYL SULFOXIDE C2 H6 O S IAZDPXIOMUYVGZ-UHFFFAOYSA-N |  | ||
| CL (Subject of Investigation/LOI) Query on CL | G [auth A], L [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| ELY Query on ELY | A, B | L-PEPTIDE LINKING | C10 H22 N2 O2 |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 46.921 | α = 74.61 |
| b = 61.804 | β = 81.74 |
| c = 75.633 | γ = 76.79 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| Aimless | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 21807088 |