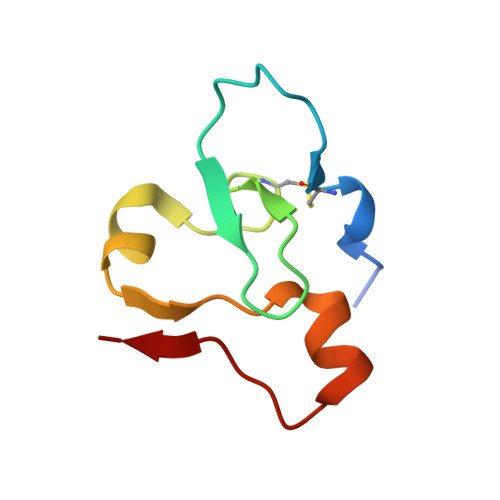
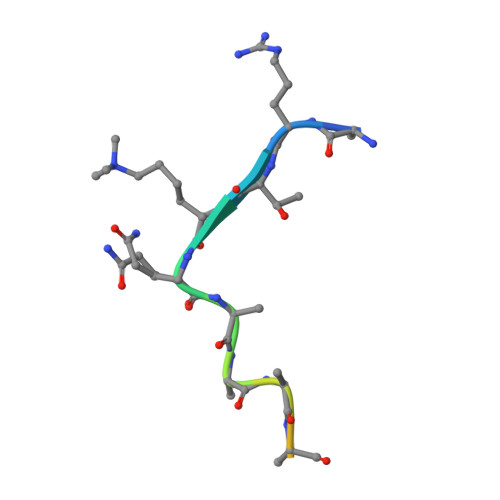
Structural insights into an atypical histone binding mechanism by a PHD finger.
Gregoire, S., Gregoire, J., Yang, Y., Capitani, S., Joshi, M., Sarvan, S., Zaker, A., Ning, Z., Figeys, D., Ulrich, K., Brunzelle, J.S., Mer, A., Couture, J.F.(2024) Structure 32: 1498
- PubMed: 39029460
- DOI: https://doi.org/10.1016/j.str.2024.06.017
- Primary Citation of Related Structures:
9C0O - PubMed Abstract:
Complex associating with SET1 (COMPASS) is a histone H3K4 tri-methyltransferase controlled by several regulatory subunits including CXXC zinc finger protein 1 (Cfp1). Prior studies established the structural underpinnings controlling H3K4me3 recognition by the PHD domain of Cfp1's yeast homolog (Spp1). However, metazoans Cfp1 PHD lacks structural elements important for H3K4me3 stabilization in Spp1, suggesting that in metazoans, Cfp1 PHD domain binds H3K4me3 differently. The structure of Cfp1 PHD in complex with H3K4me3 shows unique features such as non-canonical coordination of the first zinc atom and a disulfide bond forcing the reorientation of Cfp1 PHD N-terminus, thereby leading to an atypical H3K4me3 binding pocket. This configuration minimizes Cfp1 PHD reliance on canonical residues important for histone binding functions of other PHD domains. Cancer-related mutations in Cfp1 PHD impair H3K4me3 binding, implying a potential impact on epigenetic signaling. Our work highlights a potential diversification of PHD histone binding modes and the impact of cancer mutations on Cfp1 functions.
- Ottawa Institute of Systems Biology, Ottawa, Ontario K1H 8M5, Canada; Department of Biochemistry, Microbiology and Immunology, Faculty of Medicine, University of Ottawa, Ottawa, Ontario K1H 8M5, Canada.
Organizational Affiliation: