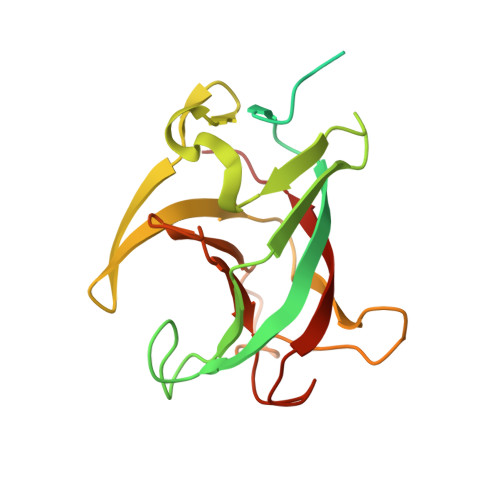
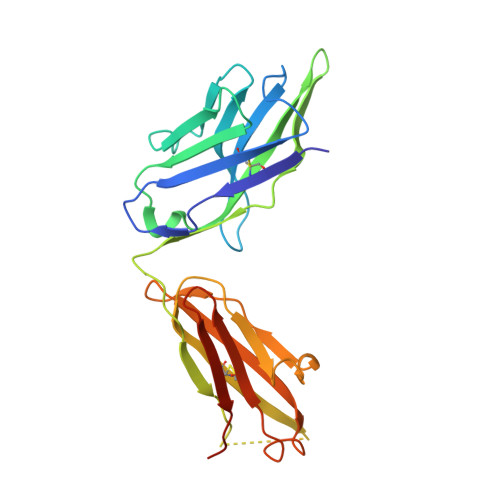
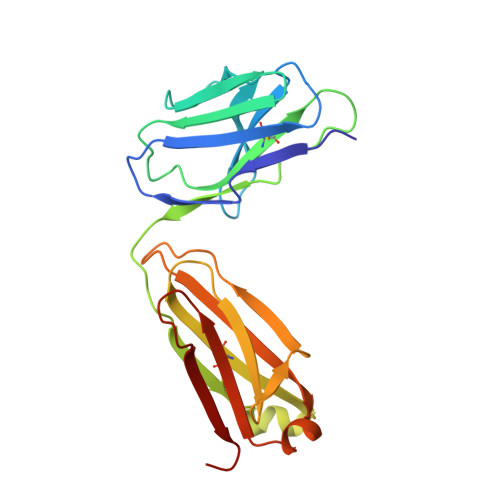
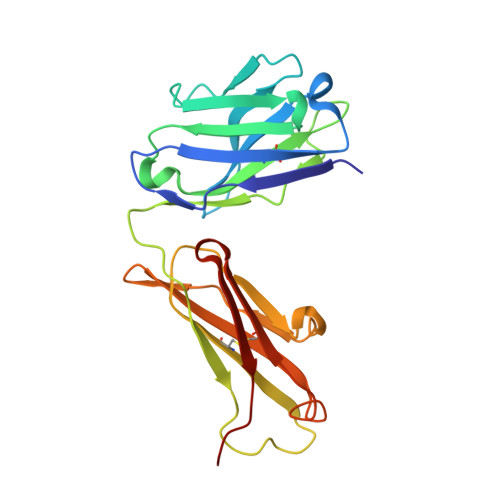
Structural basis for the synergetic neutralization of hepatitis E virus by antibody-antibody interaction.
Zheng, M., Zhou, L., Huang, Y., Zhang, X., Yu, Z., Yang, C., Chen, Y., Ying, D., Wang, H., Chen, Z., Liu, C., Tang, Z., Wang, S., Wang, K., Yang, K., Lin, Y., Li, T., Zheng, Q., Zheng, Z., Zhang, J., Yu, H., Li, S., Gu, Y., Xia, N.(2024) Proc Natl Acad Sci U S A 121: e2408585121-e2408585121
- PubMed: 39585981
- DOI: https://doi.org/10.1073/pnas.2408585121
- Primary Citation of Related Structures:
9IY0, 9IY2 - PubMed Abstract:
Neutralizing antibodies (nAbs) play a crucial role in virology, antibody drug development, and vaccine research. In this study, we investigated the synergistic effect of two hepatitis E virus (HEV) nAbs, 8H3, and 8C11, which have exhibited enhanced neutralizing activity in a rhesus monkey model. We presented crystal structures of 8H3 Fab alone and a triple complex of 8C11 Fab and 8H3 Fab simultaneously binding to the HEV E2s protein (8C11:E2s:8H3). Through structural analysis, we identified critical binding sites and fully elucidated the binding footprints of nAb 8H3 in the 8C11:E2s:8H3 complex using site-directed mutagenesis, pinpointing Ile 529, Glu 549, Lys 554, and Ser 566 in the E2s domain, and K66H, S67H, D88H in the 8C11 heavy chain. Interestingly, the synergetic enhancement of 8C11 to 8H3 converted to an antagonistic effect when 8C11 bound to E2s with pretreatment of 8H3, indicating a unidirectional synergistic effect associated with the sequence of antibody involvement. We demonstrated this phenomenon through structural comparisons of E2s:8C11 vs. 8C11:E2s:8H3 crystal structures and molecular dynamics simulations, found that Ile 529 played a key role in the synergistic interplay between these two nAbs. The two-antibody combination showed a more potent antibody-imposed physical disruption mechanism and enhanced coneutralization in an authentic HEV-based cell model. Our study suggests a strategy for synergistic antibody cocktail design with antibody-antibody side-by-side interaction.
- State Key Laboratory of Vaccines for Infectious Diseases, Xiang An Biomedicine Laboratory, Department of Laboratory Medicine, School of Public Health, School of Life Sciences, Xiamen University, Xiamen 361102, China.
Organizational Affiliation: