Structural dissection of alpha beta-tubulin heterodimer assembly and disassembly by human tubulin-specific chaperones
Seong, Y.J., Kim, H.M., Byun, K.M., Park, Y.W., Roh, S.H.(2025) Science
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2025) Science
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin-specific chaperone C | A [auth C] | 346 | Homo sapiens | Mutation(s): 0 Gene Names: TBCC | 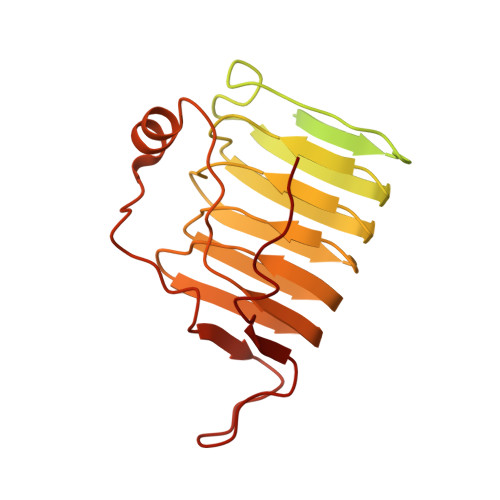 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q15814 (Homo sapiens) Explore Q15814 Go to UniProtKB: Q15814 | |||||
PHAROS: Q15814 GTEx: ENSG00000124659 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q15814 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin-specific chaperone D | B [auth D] | 1,192 | Homo sapiens | Mutation(s): 0 Gene Names: TBCD, KIAA0988, SSD1, TFCD, PP1096 | 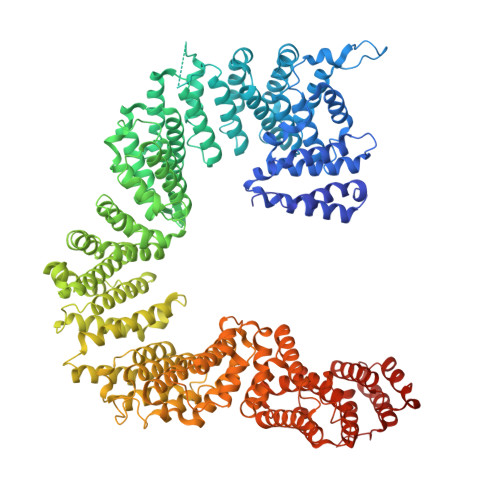 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9BTW9 (Homo sapiens) Explore Q9BTW9 Go to UniProtKB: Q9BTW9 | |||||
PHAROS: Q9BTW9 GTEx: ENSG00000141556 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9BTW9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin-specific chaperone E | C [auth E] | 527 | Homo sapiens | Mutation(s): 0 Gene Names: TBCE | 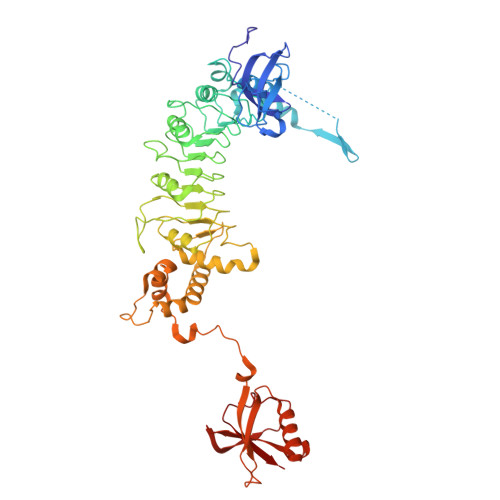 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q15813 (Homo sapiens) Explore Q15813 Go to UniProtKB: Q15813 | |||||
PHAROS: Q15813 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q15813 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ADP-ribosylation factor-like protein 2 | D [auth G] | 184 | Homo sapiens | Mutation(s): 0 Gene Names: ARL2 | 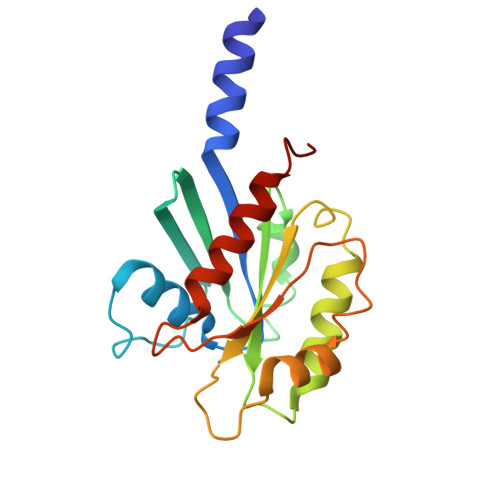 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P36404 (Homo sapiens) Explore P36404 Go to UniProtKB: P36404 | |||||
PHAROS: P36404 GTEx: ENSG00000213465 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P36404 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin alpha-1B chain | E [auth a] | 451 | Sus scrofa | Mutation(s): 0 Gene Names: TUBA1B EC: 3.6.5 |  |
UniProt | |||||
Find proteins for Q2XVP4 (Sus scrofa) Explore Q2XVP4 Go to UniProtKB: Q2XVP4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2XVP4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin beta chain | F [auth b] | 444 | Sus scrofa | Mutation(s): 0 Gene Names: TUBB, TUBB5 |  |
UniProt | |||||
Find proteins for Q767L7 (Sus scrofa) Explore Q767L7 Go to UniProtKB: Q767L7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q767L7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GTP (Subject of Investigation/LOI) Query on GTP | J [auth a], L [auth b] | GUANOSINE-5'-TRIPHOSPHATE C10 H16 N5 O14 P3 XKMLYUALXHKNFT-UUOKFMHZSA-N |  | ||
| GDP (Subject of Investigation/LOI) Query on GDP | G | GUANOSINE-5'-DIPHOSPHATE C10 H15 N5 O11 P2 QGWNDRXFNXRZMB-UUOKFMHZSA-N |  | ||
| AF3 (Subject of Investigation/LOI) Query on AF3 | H [auth G] | ALUMINUM FLUORIDE Al F3 KLZUFWVZNOTSEM-UHFFFAOYSA-K |  | ||
| MG (Subject of Investigation/LOI) Query on MG | I [auth G], K [auth a], M [auth b] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Research Foundation (NRF, Korea) | Korea, Republic Of | -- |