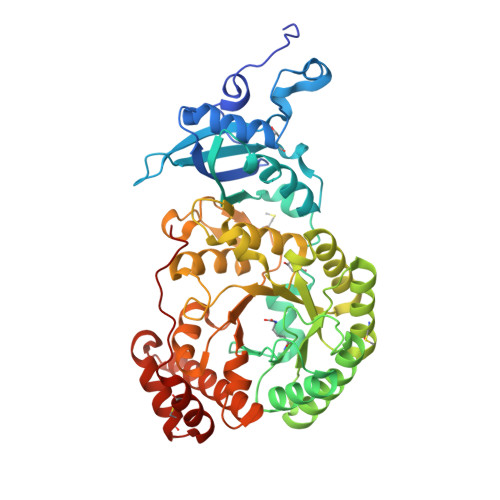
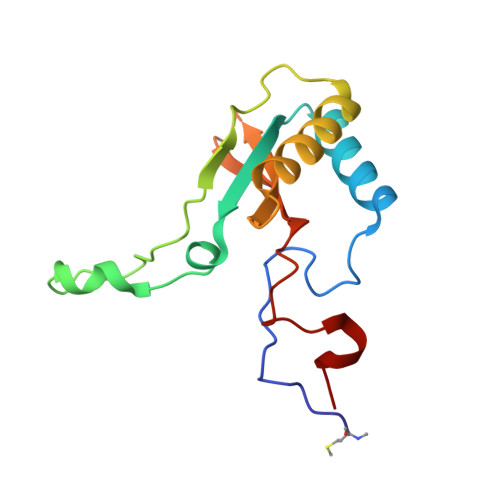
Structural and Functional Consequences of the Replacement of Proximal Residues Cys-172 and Cys-192 in the Large Subunit of Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase from Chlamydomonas Reinhardtii
Garcia-Murria, M.-J., Karkehabadi, S., Marin-Navarro, J., Satagopan, S., Andersson, I., Spreitzer, R.J., Moreno, J.(2008) Biochem J 411: 241
- PubMed: 18072944
- DOI: https://doi.org/10.1042/BJ20071422
- Primary Citation of Related Structures:
2VDH, 2VDI - PubMed Abstract:
Proximal Cys(172) and Cys(192) in the large subunit of the photosynthetic enzyme Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase; EC 4.1.1.39) are evolutionarily conserved among cyanobacteria, algae and higher plants. Mutation of Cys(172) has been shown to affect the redox properties of Rubisco in vitro and to delay the degradation of the enzyme in vivo under stress conditions. Here, we report the effect of the replacement of Cys(172) and Cys(192) by serine on the catalytic properties, thermostability and three-dimensional structure of Chlamydomonas reinhardtii Rubisco. The most striking effect of the C172S substitution was an 11% increase in the specificity factor when compared with the wild-type enzyme. The specificity factor of C192S Rubisco was not altered. The V(c) (V(max) for carboxylation) was similar to that of wild-type Rubisco in the case of the C172S enzyme, but approx. 30% lower for the C192S Rubisco. In contrast, the K(m) for CO(2) and O(2) was similar for C192S and wild-type enzymes, but distinctly higher (approximately double) for the C172S enzyme. C172S Rubisco showed a critical denaturation temperature approx. 2 degrees C lower than wild-type Rubisco and a distinctly higher denaturation rate at 55 degrees C, whereas C192S Rubisco was only slightly more sensitive to temperature denaturation than the wild-type enzyme. X-ray crystal structures reveal that the C172S mutation causes a shift of the main-chain backbone atoms of beta-strand 1 of the alpha/beta-barrel affecting a number of amino acid side chains. This may cause the exceptional catalytic features of C172S. In contrast, the C192S mutation does not produce similar structural perturbations.
Organizational Affiliation:
Departament de Bioquimica i Biologia Molecular, Facultat de Biologia, Universitat de València, Av. Dr Moliner 50, Burjassot, València E-46100, Spain.