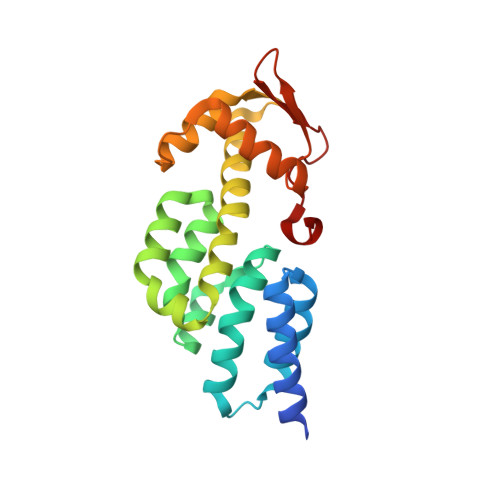
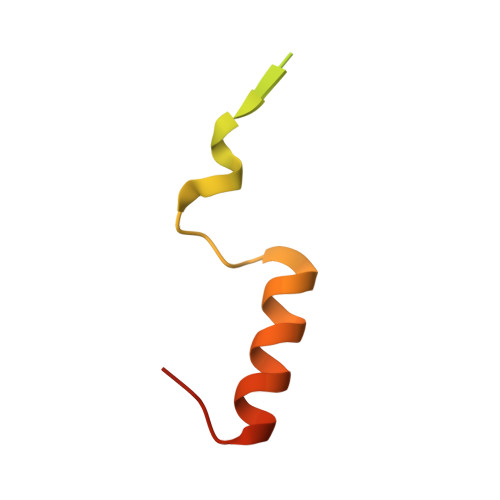
Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex.
Ellisdon, A.M., Dimitrova, L., Hurt, E., Stewart, M.(2012) Nat Struct Mol Biol 19: 328-336
- PubMed: 22343721
- DOI: https://doi.org/10.1038/nsmb.2235
- Primary Citation of Related Structures:
3T5V, 3T5X - PubMed Abstract:
The conserved TREX-2 transcription-export complex integrates transcription and processing of many actively transcribed nascent mRNAs with the recruitment of export factors at nuclear pores and also contributes to transcriptional memory and genomic stability. We report the crystal structure of the Sac3-Thp1-Sem1 segment of Saccharomyces cerevisiae TREX-2 that interfaces with the gene expression machinery. Sac3-Thp1-Sem1 forms a previously uncharacterized PCI-domain complex characterized by the juxtaposition of Sac3 and Thp1 winged helix domains, forming a platform that mediates nucleic acid binding. Our structure-guided mutations support the idea that the Thp1-Sac3 interaction is an essential requirement for mRNA binding and for the coupling of transcription and processing to mRNP assembly and export. These results provide insight into how newly synthesized transcripts are efficiently transferred from TREX-2 to the principal mRNA export factor, and they reveal how Sem1 stabilizes PCI domain-containing proteins and promotes complex assembly.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Cambridge, UK.