Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
Scott, D.E., Marsh, M., Blundell, T.L., Abell, C., Hyvonen, M.(2016) FEBS Lett 590: 1094
- PubMed: 26992456
- DOI: https://doi.org/10.1002/1873-3468.12139
- Primary Citation of Related Structures:
5FOT, 5FOU, 5FOV, 5FOW, 5FOX, 5FPK - PubMed Abstract:
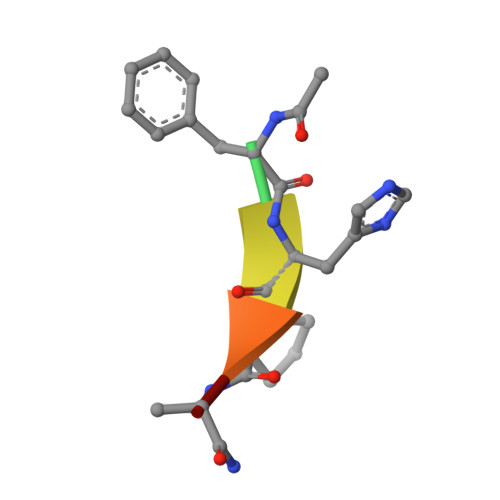
RAD51 is a recombinase involved in the homologous recombination of double-strand breaks in DNA. RAD51 forms oligomers by binding to another molecule of RAD51 via an 'FxxA' motif, and the same recognition sequence is similarly utilised to bind BRCA2. We have tabulated the effects of mutation of this sequence, across a variety of experimental methods and from relevant mutations observed in the clinic. We use mutants of a tetrapeptide sequence to probe the binding interaction, using both isothermal titration calorimetry and X-ray crystallography. Where possible, comparison between our tetrapeptide mutational study and the previously reported mutations is made, discrepancies are discussed and the importance of secondary structure in interpreting alanine scanning and mutational data of this nature is considered.
Organizational Affiliation:
Department of Chemistry, University of Cambridge, Cambridge, UK.