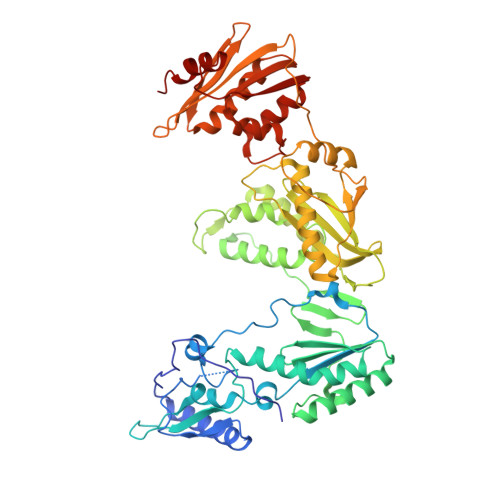
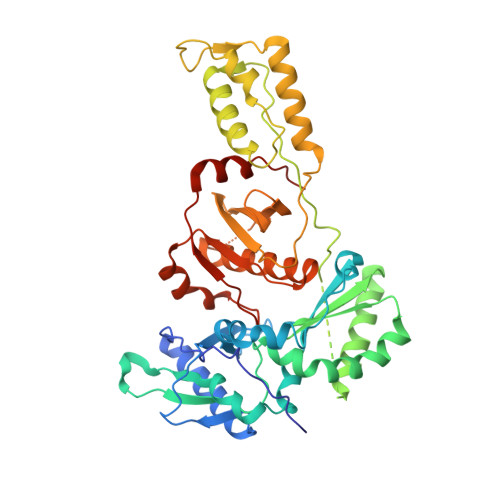
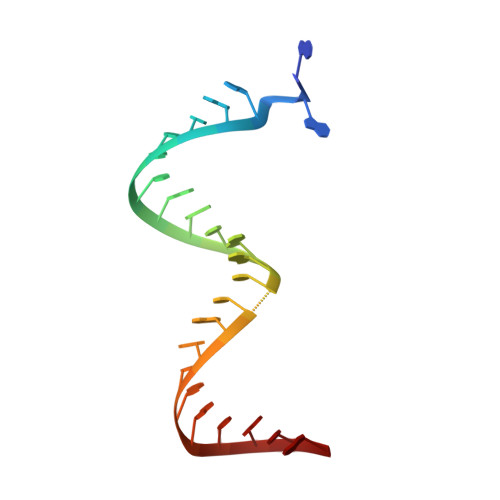
Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Tian, L., Kim, M.S., Li, H., Wang, J., Yang, W.(2018) Proc Natl Acad Sci U S A 115: 507-512
- PubMed: 29295939
- DOI: https://doi.org/10.1073/pnas.1719746115
- Primary Citation of Related Structures:
6BSG, 6BSH, 6BSI, 6BSJ - PubMed Abstract:
HIV-1 reverse transcriptase (RT) contains both DNA polymerase and RNase H activities to convert the viral genomic RNA to dsDNA in infected host cells. Here we report the 2.65-Å resolution structure of HIV-1 RT engaging in cleaving RNA in an RNA/DNA hybrid. A preferred substrate sequence is absolutely required to enable the RNA/DNA hybrid to adopt the distorted conformation needed to interact properly with the RNase H active site in RT. Substituting two nucleotides 4 bp upstream from the cleavage site results in scissile-phosphate displacement by 4 Å. We also have determined the structure of HIV-1 RT complexed with an RNase H-resistant polypurine tract sequence, which adopts a rigid structure and is accommodated outside of the nuclease active site. Based on this newly gained structural information and a virtual drug screen, we have identified an inhibitor specific for the viral RNase H but not for its cellular homologs.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892.