Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Jiang, F., Li, N., Wang, X., Cheng, J., Huang, Y., Yang, Y., Yang, J., Cai, B., Wang, Y.P., Jin, Q., Gao, N.(2019) Cell 177: 370-383.e15
- PubMed: 30905475
- DOI: https://doi.org/10.1016/j.cell.2019.02.020
- Primary Citation of Related Structures:
6J0B, 6J0C, 6J0F, 6J0M, 6J0N - PubMed Abstract:
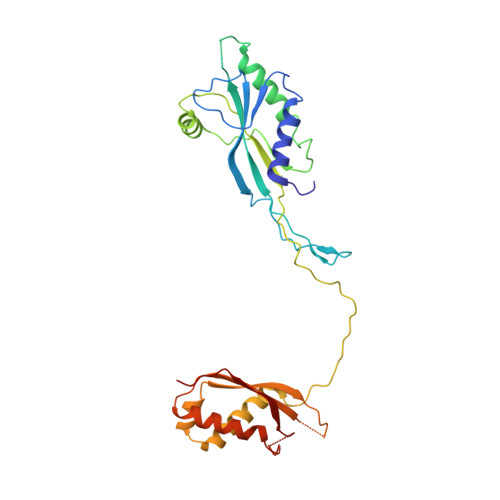
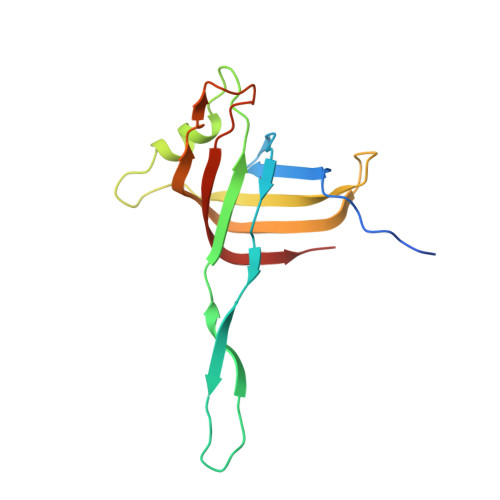
Contractile injection systems (CISs) are cell-puncturing nanodevices that share ancestry with contractile tail bacteriophages. Photorhabdus virulence cassette (PVC) represents one group of extracellular CISs that are present in both bacteria and archaea. Here, we report the cryo-EM structure of an intact PVC from P. asymbiotica. This over 10-MDa device resembles a simplified T4 phage tail, containing a hexagonal baseplate complex with six fibers and a capped 117-nanometer sheath-tube trunk. One distinct feature of the PVC is the presence of three variants for both tube and sheath proteins, indicating a functional specialization of them during evolution. The terminal hexameric cap docks onto the topmost layer of the inner tube and locks the outer sheath in pre-contraction state with six stretching arms. Our results on the PVC provide a framework for understanding the general mechanism of widespread CISs and pave the way for using them as delivery tools in biological or therapeutic applications.
Organizational Affiliation:
NHC Key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, PRC.