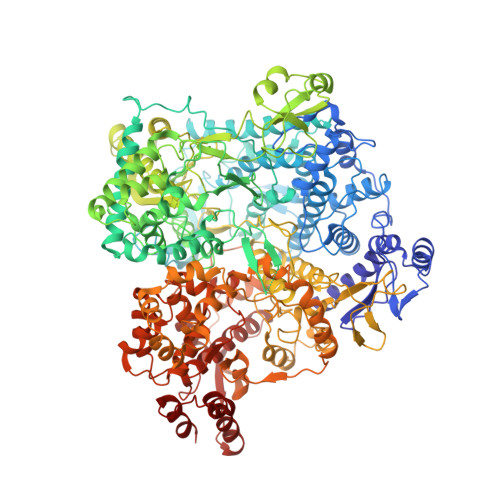
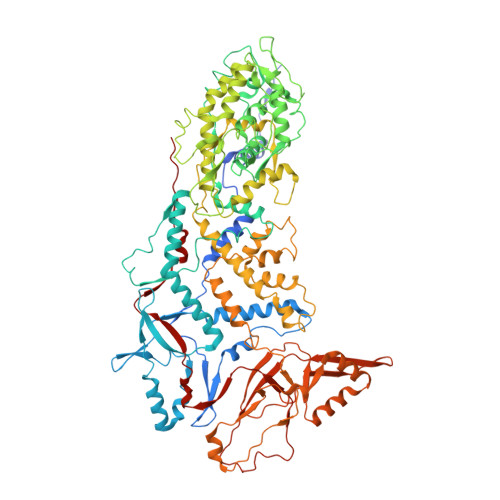
In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating.
He, Y., Shivakoti, S., Ding, K., Cui, Y., Roy, P., Zhou, Z.H.(2019) Proc Natl Acad Sci U S A 116: 16535-16540
- PubMed: 31350350
- DOI: https://doi.org/10.1073/pnas.1905849116
- Primary Citation of Related Structures:
6PNS, 6PO2 - PubMed Abstract:
Bluetongue virus (BTV), a major threat to livestock, is a multilayered, nonturreted member of the Reoviridae , a family of segmented dsRNA viruses characterized by endogenous RNA transcription through an RNA-dependent RNA polymerase (RdRp). To date, the structure of BTV RdRp has been unknown, limiting our mechanistic understanding of BTV transcription and hindering rational drug design effort targeting this essential enzyme. Here, we report the in situ structures of BTV RdRp VP1 in both the triple-layered virion and double-layered core, as determined by cryo-electron microscopy (cryoEM) and subparticle reconstruction. BTV RdRp has 2 unique motifs not found in other viral RdRps: a fingernail, attached to the conserved fingers subdomain, and a bundle of 3 helices: 1 from the palm subdomain and 2 from the N-terminal domain. BTV RdRp VP1 is anchored to the inner surface of the capsid shell via 5 asymmetrically arranged N termini of the inner capsid shell protein VP3A around the 5-fold axis. The structural changes of RdRp VP1 and associated capsid shell proteins between BTV virions and cores suggest that the detachment of the outer capsid proteins VP2 and VP5 during viral entry induces both global movements of the inner capsid shell and local conformational changes of the N-terminal latch helix (residues 34 to 51) of 1 inner capsid shell protein VP3A, priming RdRp VP1 within the capsid for transcription. Understanding this mechanism in BTV also provides general insights into RdRp activation and regulation during viral entry of other multilayered, nonturreted dsRNA viruses.
Organizational Affiliation:
Department of Microbiology, Immunology & Molecular Genetics, University of California, Los Angeles, CA 90095.