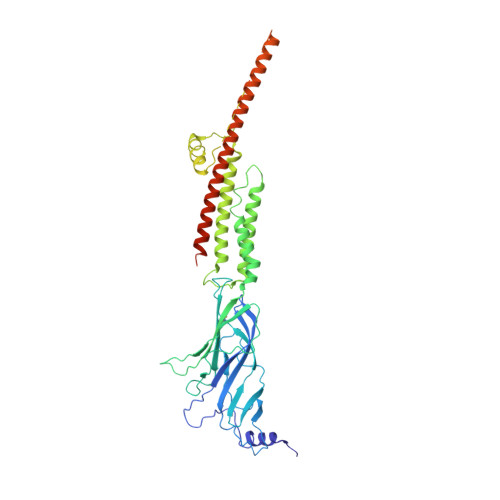
Agonist Selectivity and Ion Permeation in the alpha 3 beta 4 Ganglionic Nicotinic Receptor.
Gharpure, A., Teng, J., Zhuang, Y., Noviello, C.M., Walsh Jr., R.M., Cabuco, R., Howard, R.J., Zaveri, N.T., Lindahl, E., Hibbs, R.E.(2019) Neuron 104: 501
- PubMed: 31488329
- DOI: https://doi.org/10.1016/j.neuron.2019.07.030
- Primary Citation of Related Structures:
6PV7, 6PV8 - PubMed Abstract:
Nicotinic acetylcholine receptors are pentameric ion channels that mediate fast chemical neurotransmission. The α3β4 nicotinic receptor subtype forms the principal relay between the central and peripheral nervous systems in the autonomic ganglia. This receptor is also expressed focally in brain areas that affect reward circuits and addiction. Here, we present structures of the α3β4 nicotinic receptor in lipidic and detergent environments, using functional reconstitution to define lipids appropriate for structural analysis. The structures of the receptor in complex with nicotine, as well as the α3β4-selective ligand AT-1001, complemented by molecular dynamics, suggest principles of agonist selectivity. The structures further reveal much of the architecture of the intracellular domain, where mutagenesis experiments and simulations define residues governing ion conductance.
- Department of Neuroscience, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: