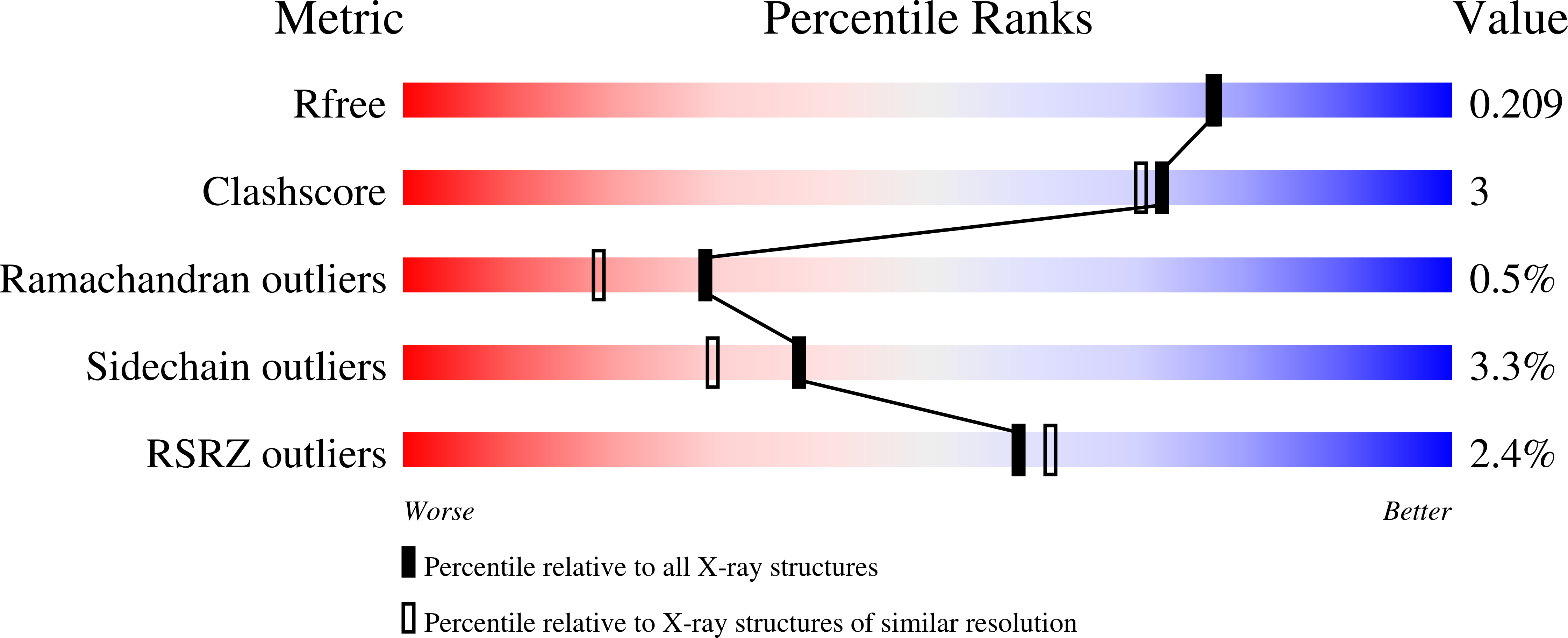
Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors (Part 2).
de Heuvel, E., Singh, A.K., Boronat, P., Kooistra, A.J., van der Meer, T., Sadek, P., Blaazer, A.R., Shaner, N.C., Bindels, D.S., Caljon, G., Maes, L., Sterk, G.J., Siderius, M., Oberholzer, M., de Esch, I.J.P., Brown, D.G., Leurs, R.(2019) Bioorg Med Chem 27: 4013-4029
- PubMed: 31378593
- DOI: https://doi.org/10.1016/j.bmc.2019.06.026
- Primary Citation of Related Structures:
6GXQ, 6HWO, 6RB6, 6RCW, 6RFN, 6RFW, 6RGK - PubMed Abstract:
Inhibitors against Trypanosoma brucei phosphodiesterase B1 (TbrPDEB1) and B2 (TbrPDEB2) have gained interest as new treatments for human African trypanosomiasis. The recently reported alkynamide tetrahydrophthalazinones, which show submicromolar activities against TbrPDEB1 and anti-T. brucei activity, have been used as starting point for the discovery of new TbrPDEB1 inhibitors. Structure-based design indicated that the alkynamide-nitrogen atom can be readily decorated, leading to the discovery of 37, a potent TbrPDEB1 inhibitor with submicromolar activities against T. brucei parasites. Furthermore, 37 is more potent against TbrPDEB1 than hPDE4 and shows no cytotoxicity on human MRC-5 cells. The crystal structures of the catalytic domain of TbrPDEB1 co-crystalized with several different alkynamides show a bidentate interaction with key-residue Gln874, but no interaction with the parasite-specific P-pocket, despite being (uniquely) a more potent inhibitor for the parasite PDE. Incubation of blood stream form trypanosomes by 37 increases intracellular cAMP levels and results in the distortion of the cell cycle and cell death, validating phosphodiesterase inhibition as mode of action.
Organizational Affiliation:
Division of Medicinal Chemistry, Amsterdam Institute for Molecules, Medicines and Systems, Vrije Universiteit Amsterdam, 1081 HZ Amsterdam, The Netherlands.