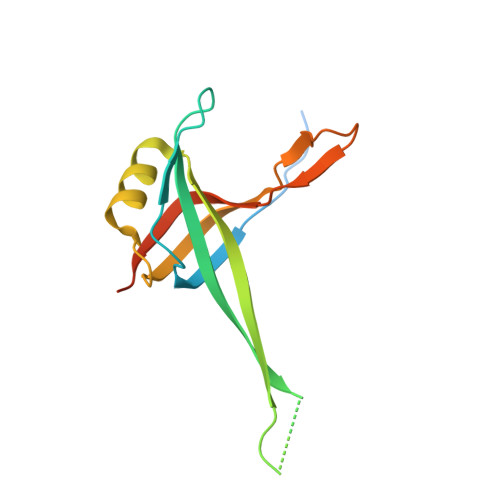
Dominant mutations in mtDNA maintenance gene SSBP1 cause optic atrophy and foveopathy.
Piro-Megy, C., Sarzi, E., Tarres-Sole, A., Pequignot, M., Hensen, F., Quiles, M., Manes, G., Chakraborty, A., Senechal, A., Bocquet, B., Cazevieille, C., Roubertie, A., Muller, A., Charif, M., Goudenege, D., Lenaers, G., Wilhelm, H., Kellner, U., Weisschuh, N., Wissinger, B., Zanlonghi, X., Hamel, C., Spelbrink, J.N., Sola, M., Delettre, C.(2020) J Clin Invest 130: 143-156
- PubMed: 31550237
- DOI: https://doi.org/10.1172/JCI128513
- Primary Citation of Related Structures:
6RUP - PubMed Abstract:
Mutations in genes encoding components of the mitochondrial DNA (mtDNA) replication machinery cause mtDNA depletion syndromes (MDSs), which associate ocular features with severe neurological syndromes. Here, we identified heterozygous missense mutations in single-strand binding protein 1 (SSBP1) in 5 unrelated families, leading to the R38Q and R107Q amino acid changes in the mitochondrial single-stranded DNA-binding protein, a crucial protein involved in mtDNA replication. All affected individuals presented optic atrophy, associated with foveopathy in half of the cases. To uncover the structural features underlying SSBP1 mutations, we determined a revised SSBP1 crystal structure. Structural analysis suggested that both mutations affect dimer interactions and presumably distort the DNA-binding region. Using patient fibroblasts, we validated that the R38Q variant destabilizes SSBP1 dimer/tetramer formation, affects mtDNA replication, and induces mtDNA depletion. Our study showing that mutations in SSBP1 cause a form of dominant optic atrophy frequently accompanied with foveopathy brings insights into mtDNA maintenance disorders.
Organizational Affiliation:
Institute of Neurosciences of Montpellier, INSERM, University of Montpellier, Montpellier, France.