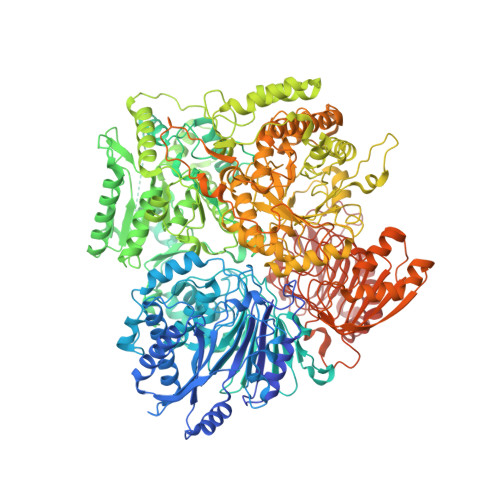
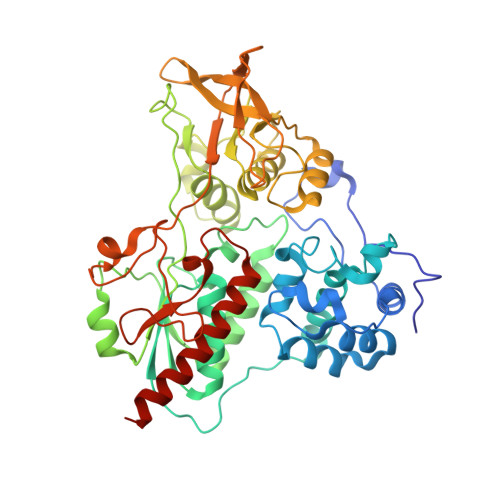
Cryo-EM Structures of Azospirillum brasilense Glutamate Synthase in Its Oligomeric Assemblies.
Swuec, P., Chaves-Sanjuan, A., Camilloni, C., Vanoni, M.A., Bolognesi, M.(2019) J Mol Biology 431: 4523-4526
- PubMed: 31473159
- DOI: https://doi.org/10.1016/j.jmb.2019.08.011
- Primary Citation of Related Structures:
6S6S, 6S6T, 6S6U, 6S6X - PubMed Abstract:
Bacterial NADPH-dependent glutamate synthase (GltS) is a complex iron-sulfur flavoprotein that catalyzes the reductive synthesis of two L-Glu molecules from L-Gln and 2-oxo-glutarate. GltS functional unit hosts an α-subunit (αGltS) and a β-subunit (βGltS) that assemble in different αβ oligomers in solution. Here, we present the cryo-electron microscopy structures of Azospirillum brasilense GltS in four different oligomeric states (α 4 β 3 , α 4 β 4 , α 6 β 4 and α 6 β 6 , in the 3.5- to 4.1-Å resolution range). Our study provides a comprehensive GltS model that details the inter-protomeric assemblies and allows unequivocal location of the FAD cofactor and of two electron transfer [4Fe-4S] +1,+2 clusters within βGltS.
Organizational Affiliation:
Dipartimento di Bioscienze, Università degli Studi di Milano, Via Celoria 26, 20133 Milano, Italy; Centro di Ricerca Pediatrica Romeo ed Enrica Invernizzi, Università degli Studi di Milano, Via Celoria 26, 20133 Milano, Italy.